p16 expression correlates with basal-like triple-negative breast carcinoma
Amany A Abou-Bakr 1 and Hany I Eldweny 2
1 Department of Pathology, National Cancer Institute, Cairo University, Egypt
2 Department of Surgery, National Cancer Institute, Cairo University, Egypt
Correspondence to: Amany A Abou-Bakr. Email: amany_aboubakr@hotmail.com
Abstract
Background: Basal-like breast carcinoma (BLBC) has attracted considerable attention over the past few years. It has been suggested that tumours expressing basal markers have a more aggressive clinical behaviour. However, a molecular basis for this disease remains unclear, and it lacks currently used therapeutic targets. Therefore developing a novel treatment strategy is crucial for improving the prognosis. The aim of this study was to characterise the immunohistochemical (IHC) expression of p16 in patients with BLBC compared with non-BLBC.
Materials and methods: Eighty-five cases of grade-3 invasive ductal carcinomas not otherwise specified (IDC-NOS) were analyzed. Immunohistochemical stains for oestrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor type 2 (HER2), cytokeratin (CK) 5/6, epidermal growth factor receptor (EGFR) and p16 were performed. BLBC was defined as ER-, PR-, Her2- and CK5/6 , and/or EGFR .
Results: Twenty cases were categorised as BLBC versus 65 as non-basal. High mitotic count and presence of necrosis were associated with basal-like phenotype. Distant metastasis developed in 40% of cases of BLBC with frequent spread to brain and lung. p16 had significantly higher expression in the basal subgroup (80% versus 50.8%, P = 0.04). Patients with BLBCs were found to have a lower disease-free survival (DFS) rate (60% versus 70.8%, P = 0.03).
Conclusion: BLBC typically demonstrates a unique profile. p16 is frequently expressed in breast cancers with basal-like phenotype; this suggests that p16 may play a role in the poor prognosis of this tumour, and it may be used in the development of a targeted therapy that will result in improved patient prognostication and outcome.
Keywords: Basal-like, breast cancer, p16.
Introduction
Breast carcinomas are considered to be a heterogeneous group of tumours showing different behaviour, prognosis, and response to treatment [1]. Furthermore, tumours classified under the same histological type and grade can present distinct molecular aspects and biological course. The molecular heterogeneity of breast tumours cannot be morphologically assessed, and it represents an important challenge for the research and treatment of breast cancer [2]. Recently, gene expression profiling analyses using DNA microarrays and later on immunohistochemical (IHC) studies have enabled the recognition of distinct subtypes of tumours associated with different clinical outcomes [3–5]: luminal A (positive for oestrogen receptor (ER) and progesterone receptor (PR) and negative for human epidermal growth factor receptor type 2 [HER2]); luminal B (ER , PR , HER2 ); HER2 overexpressing (ER-, PR-, HER2 ); basal-like (differentiation towards basal cell types); unclassifiable or normal breast-like (tumours that are negative for all the markers above).
There is no consensus on how to define basal-like breast carcinoma (BLBC) based on immunohistochemistry. The majority of BLBCs lack the expression of ER, PR, and HER2 protein overexpression [4, 6, 7]. BLBC has also been characterised by the expression of cytokeratin (CK) 5/6 and 17, epidermal growth factor receptor (EGFR), c-kit, and vascular endothelial growth factor [7, 8, 9]. Nielsen et al [7] have developed an IHC panel for identifying BLBCs on the basis of a comparison between the transcriptomic and IHC profiles. According to this definition, BLBCs are negative for ER and HER2 and positive for CK5/6 and/or EGFR. Conversely, others have proposed that a proportion of BLBCs may be positive for ER and HER2 [10, 11].
The prevalence of BLBC ranges from 7.5% to 36.7% of breast cancer cases in different patient cohorts [3, 5, 7, 12–23]. These tumours were predominantly grade-3, displaying a high mitotic count, tumour necrosis, pushing margin of invasion, stromal lymphocytic response, high rates of nuclear pleomorphism, and a lack of tubule formation [24–26].
Basal-like breast cancer remains a great challenge because of its clinically aggressive nature and poorly characterised molecular pathogenesis. Unlike ER-positive luminal tumours and HER2-positive tumours, the basal-like subtype typically lacks expression of the molecular targets that confer responsiveness to highly effective targeted therapies such as tamoxifen, aromatase inhibitors, or trastuzumab. Indeed, identification of the relevant molecular targets in BLBC remains a formidable challenge.
The cyclin-dependent kinase inhibitor-2A gene (p16INK4a), located within the CDKN2A locus, is a strong and specific inhibitor of the progression through the G1 phase of the cell cycle by preventing phosphorylation of Rb protein [27] and is considered a major tumour suppressor gene. The inactivation of p16 seems a crucial event in the development of several human tumours [28]. Hui et al [29] demonstrated an inverse relationship between p16INK4a and ER mRNA levels in cell lines and primary breast cancers, suggesting that p16INK4a inactivation by hypermethylation and overexpression is a marker of poor prognosis. Similarly, Milde-Langosch et al [30] have described high p16INK4a reactivity (both nuclear and cytoplasmic) as indicative of a more undifferentiated phenotype in mammary carcinomas.
The aim of this study was to characterise the IHC expression of p16 in patients with BLBC compared with non-BLBC in a group of grade-3 invasive ductal carcinoma not otherwise specified (IDC-NOS) to identify a more homogenous subset to which targeted therapy can be applied.
Material and methods
Tissue samples
Eighty-five randomly selected cases that underwent surgical treatment and reported as grade-3 IDC-NOS (a subgroup with a traditionally poor prognosis) were analysed in this work. The study protocol was in accordance with the ethical guidelines of the 2004 Declaration of Helsinki. Patients with primary tumours for which paraffin blocks and follow-up data were available were retrieved from the Department of Surgical Pathology, National Cancer Institute, Cairo University, Egypt between January 2000 and January 2005. Special histologic subtypes were not included to avoid any confounding effect, and none of the patients had received adjuvant therapy before surgery. Tumour morphologic parameters including mitotic activity, necrosis, and Nottingham grade [31] were evaluated.
Immunohistochemistry
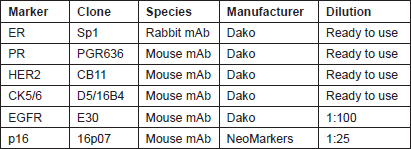
Four micron tissue sections were deparaffinised in xylene and rehydrated through a series of decreasing ethanol concentration. The slides were pretreated with hydrogen peroxide (3%) for 10 minutes to remove the endogenous peroxidase, followed by antigen retrieval in microwave for 15 minutes in 10 mM citrate buffer (pH 6.0). The primary antibodies were applied, followed by washing and incubation with the biotinylated secondary antibody for 30 minutes at room temperature. The slides were counterstained with haematoxylin and dehydrated in alcohol and xylene before mounting. Appropriate positive and negative controls were included with each IHC run. The characteristics of the primary antibodies used in this study are listed in Table 1.
Table 1: Characteristics of the primary antibodies.
Evaluation of IHC Staining
Tumours were considered to be positive for ER and PR when nuclear positivity was observed in >1% of neoplastic cells [32]. HER2 scoring was performed according to the UK accepted diagnosis criteria [33]. Patients with scores of 0 and 1 were considered negative, whereas 3 positive. Equivocal cases (2 ) were referred for fluorescence in situ hybridisation (FISH) using the Vysis PathVysion HER2 DNA probe kit (Abbott Molecular, Inc, Abbott Park, IL) [34].
For CK5/6, and EGFR > 10% cytoplasmic and/or membranous staining was considered positive [35]. Cases were recorded positive for p16 based on the presence of nuclear and/or cytoplasmic reactivity in >10% of tumour cells [36] with strong intensity. p16 evaluation was done blinded to BLBC category.
In this study, we defined BLBC using the criteria of Carey et al and others by the negativity to ER, PR, and HER2 plus the expression CK5/6 and/or EGFR [7, 13, 37].
Statistical Analysis
Statistical analysis was performed using the computer software StatView (Abacus Concepts, Inc.). Chi-square and Fisher exact tests were used to study the statistical association between categorical variables. Unpaired t test was used for analysis of continuous variables. Disease-free survival (DFS) times were estimated in months from the date of diagnosis to the date of relapse, death, or last follow-up. Survival curves were plotted using the Kaplan–Meier method, and differences in survival curves were assessed by the log-rank test. Statistical significance was defined as a P < 0.05.
Results
Of the 85 cases, 20 had a basal-like breast cancer profile performed (Figure 1). They occurred at a slightly, but significantly, younger age than other grade-3 tumours. Although we found a higher proportion of premenopausal women (65%) in the basal-like phenotype, this difference was not statistically significant (P = 0.46).
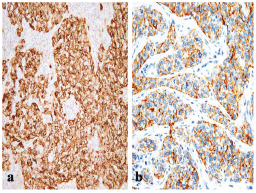
Figure 1: A BLBC that is positive for (a) EGFR (x200) and for (b) CK5/6 (x400).
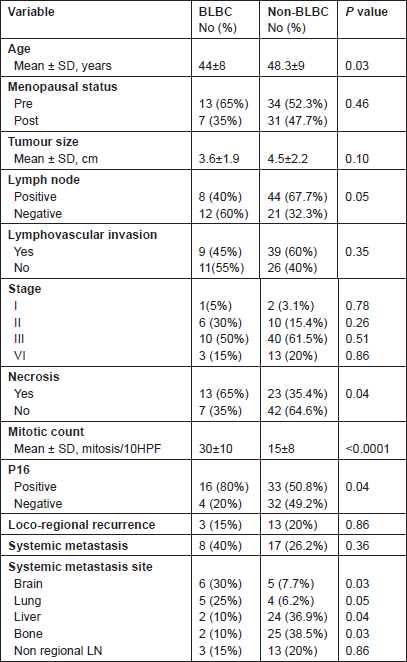
There were no statistically significant differences regarding primary tumour size, presence of vascular invasion and tumour stage (P > 0.05). BLBC was less likely to have axillary nodal metastasis at diagnosis (P = 0.05).
The presence of necrosis and mitotic count (Figure 2) showed differences between the two groups. Necrosis was more prevalent in the basal-like tumour group compared with the non-basal group (65% versus 35.4%, P = 0.04). A difference was also apparent between the mean mitotic counts, the non-basal mean (15 mitosis/10 high-power fields [HPF], range 8–50) was significantly lower than that of the basal-like group (30 mitosis/10 HPF, range 10–70).
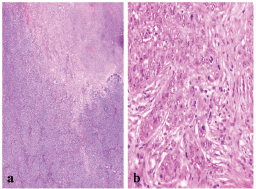
Figure 2: A BLBC showing (a) central necrosis (x200), and (b) sheets of markedly pleomorphic cells with conspicuous mitosis (x400).
The frequency of p16 positive cases was higher in the BLBC (Figure 3). p16 was expressed in 16/20 (80%) in basal cancers and in 33/65 (50.8%) of non-BLBC with a significant difference (P = 0.04). In the basal category, positive cases demonstrated labelling in >75% of neoplastic cells with a mean of 81%, while the mean was 32% (range 10%–90%) in non-basal tumours.
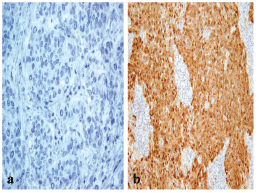
Figure 3: A p16 stain revealing (a) negative reaction in non-BLBC (x400) and (b) diffuse cytoplasmic/nuclear positivity in BLBC (x200).
Follow-up, outcome and sites of metastases
Average time of follow-up was 37.7 months (ranging from six to 60 months). In the whole cohort, 41 patients developed loco-regional or distant relapse, among these 23 patients died of breast cancer.
In BLBC, local recurrence was detected in three cases (15%), whereas systemic relapse was reported in 8/20 patients (40%), brain and lungs being the main sites (30% and 25%, respectively). Other sites of metastases were liver (10%), bone (10%) and non-regional lymph nodes (15%). There were clear differences between the basal and non-basal tumours in relation to distant metastatic sites (Table 2). Basal tumours were more likely to develop brain and lung metastases (P = 0.03 and 0.05, respectively) but significantly less likely to develop bone (P = 0.03) or liver (P = 0.04). There was no difference in the rate of involvement of non-regional lymph nodes (P = 0.86).
Table 2: Clinicopathologic characteristics of the studied cases.
After performing a log-rank test, survival analyses showed that patients with BLBC had a worse DFS when compared with patients with non-basal tumours (60% versus 70.8%, P = 0.03) (Figure 4).
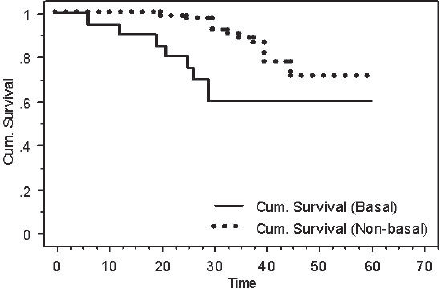
Figure 4: Kaplan-Meier DFS for basal versus non-basal tumours (P = 0.03).
Discussion
Basal-like tumours are gaining an increasing amount of attention in part owing to recognition as a distinct entity, but most importantly owing to the overall poor prognosis that the diagnosis indicates. In this study, BLBC was associated with shorter DFS. Most gene profiling studies have repeatedly reported a shorter metastasis free and overall survival among basal breast cancer patients [3, 12, 14, 16, 17, 38–42]. According to three different multigene expression signatures, most of the tumours predicted as poor prognosis were basal [43]. Data are variable with IHC [42], possibly because the terminology and definitions surrounding the concept of basal tumours are controversial, and a plethora of different markers have been employed to identify cases in clinical studies.
Also our results revealed that approximately 40% of basal-like carcinomas developed distant metastasis, more often to brain and lung than to the liver or bone [44–49]. These findings suggest that basal-like tumours might possess a distinct mechanism of metastatic spread. In fact, our observations together with the absence of association with lymph node involvement, or loco-regional relapse do not appear to justify a more radical approach to local or axillary surgery. The potentially aggressive behavior of these tumours may be better approached by the development of new systemic therapeutic strategies and targeting molecular alterations. Recent clinical trials are currently focusing on identifying these possible targets. Therefore, the main objective of this study was to examine p16 expression in basal phenotype to help in defining molecular features of this breast cancer subset.
p16 protein overexpression has been shown to be associated with breast carcinomas having poor prognostic factors [29, 30]. Importantly, virtually all of these studies occurred before the entity of basal-like cancer was established by gene expression profiling in 2001. Since the recognition of this entity, the p16 status of BLBC has not been systematically assessed. One study [50] indicated that BLBC associated with BRCA1 gene inactivation expresses lower levels of p16 than carcinomas associated with inactivation of BRCA2, which were more frequently ER positive. In addition, a review of BLBC- and BRCA1-associated tumours also states that these tumours are characterised by lower levels of p16 than the typical breast carcinoma [51]. In contrast, Gauthier et al [52] in the setting of a study of ductal carcinoma in situ noticed that BLBC show high levels of p16 transcripts and low levels of Rb transcripts. Analysing a subset of primary tumours representing each of the five molecular subtypes of breast cancer, they showed that p16 IHC labelling correlated well with mRNA levels in BLBC, although they did not statistically compare their p16 results in BLBC with those of the other different molecular subtypes. They hypothesised that loss of functional p16/Rb signalling may play a defining role in the biology of this tumour subtype. Our results correlate with their hypothesis where BLBC demonstrated significantly higher frequency of p16 positivity compared with non-BLBC.
Similarly Rakha et al’s results [21] support this and showed a difference in the expression of cell cycle regulators between basal and non-basal triple-negative tumours with p16 protein being expressed at high levels by BLBC. In accordance Bohn et al [53] found a strong positive reaction with p16 antibody in 16 out of 18 (89%) basal cases. They propose p16 as a biomarker for identification of truly basal-like cancers and raise the possibility that triple-negative breast cancer with basal cytokeratin, and p16 co-expression may adequately identify these tumours and serve as a potential diagnostic/prognostic biomarker.
Subhawong et al [54] examined the Rb/p16 pathway in BLBC because of their distinct morphologic similarities to human papilloma virus (HPV)-related poorly differentiated squamous cell carcinomas (SCCs). On the basis of similar morphology, they hypothesised that similar genes may be inactivated in these neoplasms, albeit by different mechanisms (genomic or epigenetic alterations in breast cancer, HPV in SCCs). Their data demonstrated Rb-/p16 immunophenotype in 15 of 21 BLBCs and 9 of 12 unclassifiable triple-negative carcinomas, but only 1 of 14 HER2-positive cases and none of the 17 luminal A or 7 luminal B cases (P < 0.01). So they suggested that BLBC represents a non-HPV-related carcinoma in which basal-like morphology predicts inactivation of Rb protein and diffuse p16 expression.
Another intriguing link between p16INK4a and basal-like cells comes from studies on human mammary epithelial cells, which have been shown to resemble the basal-like subtype by gene expression analysis [55, 56]. It was suggested that RB1 recruits Polycomb repression complexes to the p16INK4a locus, which silence p16INK4a transcription [57].
In summary, we suggest that p16 may define a more homogenous subgroup of BLBC, which allow further stratification of these tumours enabling more efficacious therapeutic strategy for individual patients and lessen the burden of over treatment.
Conclusion
BLBC are a distinct clinical and pathological group representing about 25% of grade-3 invasive ductal carcinomas. They show a specific pattern of distant metastasis with an increased propensity for metastases to brain and lung, sites known to be associated with a poorer prognosis. p16 is a frequent occurrence in these cancers and may play a role in the poor outcome; therefore it might be used as a potential therapeutic target to alter the clinical course of the disease and improve the survival for individual patients. Further studies on a larger cohort are needed to confirm these findings.
References
1. Payne SJ, Bowen RL, Jones JL and Wells CA (2008) Predictive markers in breast cancer - the present Histopathology 52 82–90 DOI: 10.1111/j.1365-2559.2007.02897.x PMID: 18171419
2. Rakha EA, El-Sayed ME, Reis-Filho JS and Ellis IO (2008) Expression profiling technology: its contribution to our understanding of breast cancer Histopathology 52 67–81 DOI: 10.1111/j.1365-2559.2007.02894.x PMID: 18171418
3. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE and Børresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumour subclasses with clinical implications Proc Natl Acad Sci USA 98 10869–74 DOI: 10.1073/pnas.191367098 PMCID: 58566
4. Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW and Ellis IO (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterized series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses Int J Cancer 116 340–50 DOI: 10.1002/ijc.21004 PMID: 15818618
5. Munjal K, Ambaye A, Evans M, Mitchell J, Nandedkar S and Cooper K (2009) Imunohistochemical analysis of ER, PR, Her2 and CK5/6 in infiltrative breast carcinomas in Indian patients Asian Pacific J Cancer Prev 10 773–8 PMID: 20104967
6. Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI and Ellis IO (2004) Expression of luminal and basal cytokeratins in human breast carcinoma J Pathol 203 661–71 DOI: 10.1002/path.1559 PMID: 15141381
7. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M and Perou CM (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma Clin Cancer Res 10 5367–74 DOI: 10.1158/1078-0432.CCR-04-0220 PMID: 15328174
8. Brenton JD, Carey LA, Ahmed AA and Caldas C (2005) Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 23 7350–60 DOI: 10.1200/JCO.2005.03.3845 PMID: 16145060
9. Linderholm BK, Hellborg H, Johansson U, Elmberger G, Skoog L, Lehtiö J and Lewensohn R (2009) Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer Ann Oncol 20 1639–46 DOI: 10.1093/annonc/mdp062 PMID: 19549711
10. Matos I, Dufloth R, Alvarenga M, Zeferino LC and Schmitt F (2005) p63, cytokeratin 5, and P-cadherin: three molecular markers to distinguish basal phenotype in breast carcinomas Virchows Arch 447 688–94 DOI: 10.1007/s00428-005-0010-7 PMID: 16012853
11. Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, Morandi P, Fan C, Rabiul I, Ross JS, Hortobagyi GN and Pusztai L (2005) Breast cancer molecular subtypes respond differently to preoperative chemotherapy Clin Cancer Res 11 5678–85 DOI: 10.1158/1078-0432.CCR-04-2421 PMID: 16115903
12. Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL and Botstein D (2003) Repeated observation of breast tumour subtypes in independent gene expression data sets Proc Natl Acad Sci USA 100 8418–23 DOI: 10.1073/pnas.0932692100 PMID: 12829800
13. Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS and Millikan RC (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study JAMA 295 2492–502 DOI: 10.1001/jama.295.21.2492 PMID: 16757721
14. Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS and Perou CM (2006) The molecular portraits of breast tumour are conserved across microarray platforms BMC Genomics 7 96 DOI: 10.1186/1471-2164-7-96 PMID: 16643655 PMCID: 1468408
15. Kim MJ, Ro JY, Ahn SH, Kim HH, Kim SB and Gong G (2006) Clinicopathologic significance of the basal-like subtype of breast cancer: a comparison with hormone receptor and Her2/neu-overexpressing phenotypes Hum Pathol 37 1217–26 DOI: 10.1016/j.humpath.2006.04.015 PMID: 16938528
16. Perreard L, Fan C, Quackenbush JF, Mullins M, Gauthier NP, Nelson E, Mone M, Hansen H, Buys SS, Rasmussen K, Orrico AR, Dreher D, Walters R, Parker J, Hu Z, He X, Palazzo JP, Olopade OI, Szabo A, Perou CM and Bernard PS (2006) Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay Breast Cancer Res 8 R23 DOI: 10.1186/bcr1399 PMID: 16626501 PMCID: 1557722
17. Langerød A, Zhao H, Borgan Ø, Nesland JM, Bukholm IR, Ikdahl T, Kåresen R, Børresen-Dale AL and Jeffrey SS (2007) TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer Breast Cancer Res 9 R30 DOI: 10.1186/bcr1675 PMCID: 1929092
18. Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, Hewitt SM, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R and García-Closas M (2007) Differences in risk factors for breast cancer molecular subtypes in a population-based study Cancer Epidemiol Biomarkers Prev 16 439–43 DOI: 10.1158/1055-9965.EPI-06-0806 PMID: 17372238
19. Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT, Jackson S, Nyante S, Livasy C, Carey L, Earp HS and Perou CM (2008) Epidemiology of basal-like breast cancer Breast Cancer Res Treat 109 123–39 DOI: 10.1007/s10549-007-9632-6 PMCID: 2443103
20. Huo D, Ikpatt F, Khramtsov A, Dangou JM, Nanda R, Dignam J, Zhang B, Grushko T, Zhang C, Oluwasola O, Malaka D, Malami S, Odetunde A, Adeoye AO, Iyare F, Falusi A, Perou CM and Olopade OI (2009) Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer J Clin Oncol 27 4515–21 DOI: 10.1200/JCO.2008.19.6873 PMID: 19704069 PMCID: 2754904
21. Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA, Evans AJ, Blamey R, Reis-Filho JS, Foulkes WD and Ellis IO (2009) Triple-negative breast cancer: distinguishing between basal and non basal subtypes Clin Cancer Res 15 2302–10 DOI: 10.1158/1078-0432.CCR-08-2132 PMID: 19318481
22. Toft DJ and Cryns VL (2011) Minireview: basal-like breast cancer: from molecular profiles to targeted therapies Mol Endocrinol 25 199–211 DOI: 10.1210/me.2010-0164 PMCID: 3035993
23. Cakir A, Gonul I and Uluoglu O (2012) A comprehensive morphological study for basal-like breast carcinomas with comparison to non basal-like carcinomas Diagnostic Pathol 7 145 DOI: 10.1186/1746-1596-7-145
24. Fulford LG, Easton DF, Reis-Filho JS, Sofronis A, Gillett CE, Lakhani SR and Hanby A (2006) Specific morphological features predictive for the basal phenotype in grade-3 invasive ductal carcinoma of breast Histopathology 49 22–34 DOI: 10.1111/j.13652559.2006.02453.x PMID: 16842243
25. Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT and Perou CM (2006) Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma Mod Pathol 19 264–71 DOI: 10.1038/modpathol.3800528
26. Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF and Ellis IO (2006) Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation J Pathol 208 495–506 DOI: 10.1002/path.1916 PMID: 16429394
27. Sherr CJ (1996) Cancer cell cycles Science 274 1672–77 DOI: 10.1126/science.274.5293.1672 PMID: 8939849
28. Ruas M and Peters G (1998) The p16INK4a/CDKN2A tumour suppressor and its relatives Biochim Biophys Acta 1378 115–77 PMID: 9823374
29. Hui R, Macmillan RD, Kenny FS, Musgrove EA, Blamey RW, Nicholson RI, Robertson JF and Sutherland RL (2000) INK4a gene expression and methylation in primary breast cancer: overexpression of p16INK4a messenger RNA is a marker of poor prognosis Clin Cancer Res 6 2777–87 PMID: 10914724
30. Milde-Langosch K, Bamberger AM, Rieck G, Kelp B and Loning T (2001) Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype Breast Cancer Res Treat 67 61–70 DOI: 10.1023/A:1010623308275 PMID: 11518467
31. Elston CW and Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up Histopathology 19 403–10 DOI: 10.1111/j.1365-2559.1991.tb00229.x PMID: 1757079
32. Fitzgibbons PL, Murphy DA, Hammond ME, Allred DC and Valenstein PN (2010) Recommendations for validating estrogen and progesterone receptor immunohistochemistry assays Arch Pathol Lab Med 134 930–5 PMID: 20524870
33. Hsi ED and Tubbs RR (2004) Guidelines for HER2 testing in the UK J Clin Pathol 57 241–2 DOI: 10.1136/jcp.2003.009308 PMID: 14990591
34. Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM and Hayes DF (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer J Clin Oncol 25 118–45 DOI: 10.1200/JCO.2006.09.2775
35. De Brot M, Soares FA, Stiepcich MM, Cúrcio VS and Gobbi H (2009) Basal-like breast cancers: clinicopathological features and evolutive profile Rev Assoc Med Bras 55 529–34 DOI: 10.1590/S0104-42302009000500014 PMID: 19918651
36. Chae S, Sohn J, Kim D-H, Choi YJ, Park YL, Kim K, Cho YH, Pyo JS and Kim JH (2011) Overexpression of cyclin B1, cdc2, p16, and p53 in human breast cancer: the clinicopathologic correlations and prognostic implications Yonsei Med J 52 445–53 DOI: 10.3349/ymj.2011.52.3.445 PMID: 21488187 PMCID: 3101063
37. Spitale A, Mazzola P, Soldini D, Mazzucchelli L and Bordoni A (2009) Breast cancer classification according to immunohistochemical markers: clinicopathologic features and short-term survival analysis in a population-based study from the South of Switzerland.Ann Oncol 20 628–35 DOI: 10.1093/annonc/mdn675
38. Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL and Liu ET (2003) Breast cancer classification and prognosis based on gene expression profiles from a population-based study Proc Natl Acad Sci USA 100 10393–8 DOI: 10.1073/pnas.1732912100 PMID: 12917485
39. Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Cervera N, Tarpin C, Nguyen C, Xerri L, Houlgatte R, Jacquemier J, Viens P and Birnbaum D (2005) Gene expression profiling identifies molecular subtypes of inflammatory breast cancer Cancer Res 65 2170–78 DOI: 10.1158/0008-5472.CAN-04-4115 PMID: 15781628
40. Calza S, Hall P, Auer G, Bjöhle J, Klaar S, Kronenwett U, Liu ET, Miller L, Ploner A, Smeds J, Bergh J and Pawitan Y (2006) Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients Breast Cancer Res 8 R34 DOI: 10.1186/bcr1517
41. Bertucci F, Finetti P, Cervera N, Charafe-Jauffret E, Buttarelli M, Jacquemier J, Chaffanet M, Maraninchi D, Viens P and Birnbaum D (2009) How different are luminal A and basal breast cancers? Int J Cancer 124 1338–48 DOI: 10.1002/ijc.24055
42. Bertucci F, Finetti P and Birnbaum D (2012) Basal breast cancer: A complex and deadly molecular subtype Curr Mol Med 12 96–110 DOI: 10.2174/156652412798376134 PMCID: 3343384
43. Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ and Perou CM (2006) Concordance among gene expression-based predictors for breast cancer N Engl J Med 355 560–9 DOI: 10.1056/NEJMoa052933 PMID: 16899776
44. Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN and Pusztai L (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer J Clin Oncol 26 1275–81 DOI: 10.1200/JCO.2007.14.4147 PMID: 18250347
45. Tsuda H, Takarabe T, Hasegawa F, Fukutomi T and Hirohashi S (2000) Large central acellular zones indicating myoepithelial tumour differentiation in high-grade invasive ductal carcinomas as markers of predisposition to lung and brain metastases Am J Surg Pathol 24 197–202 DOI: 10.1097/00000478-200002000-00005 PMID: 10680887
46. Hicks DG, Short SM, Prescott NL, Tarr SM, Coleman KA, Yoder BJ, Crowe JP, Choueiri TK, Dawson AE, Budd GT, Tubbs RR, Casey G and Weil RJ (2006) Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/6, and overexpress HER2 or EGFR Am J Surg Pathol 30 1097–104 DOI: 10.1097/01.pas.0000213306.05811.b9 PMID: 16931954
47. Fulford LG, Reis-Filho JS, Ryder K, Jones C, Gillett CE, Hanby A, Easton D and Lakhani SR (2007) Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival Breast Cancer Res 9 R4 DOI: 10.1186/bcr1636 PMCID: 1851397
48. Crabb SJ, Cheang MC, Leung S, Immonen T, Nielsen TO, Huntsman DD, Bajdik CD and Chia SK (2008) Basal breast cancer molecular subtype predicts for lower incidence of axillary lymph node metastases in primary breast cancer Clin Breast Cancer 8 249–56 DOI: 10.3816/CBC.2008.n.028 PMID: 18650155
49. Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA and Martens JW (2008) Subtypes of breast cancer show preferential site of relapse Cancer Res 68 3108–14 DOI: 10.1158/0008-5472.CAN-07-5644 PMID: 18451135
50. Palacios J, Honrado E, Osorio A, Cazorla A, Sarrió D, Barroso A, Rodríguez S, Cigudosa JC, Diez O, Alonso C, Lerma E, Dopazo J, Rivas C and Benítez J (2005) Phenotypic characterization of BRCA1 and BRCA2 tumours based in a tissue microarray study with 37 immunohistochemical markers Breast Cancer Res Treat 90 5–14 DOI: 10.1007/s10549-004-1536-0 PMID: 15770521
51. Turner NC and Reis-Filho JS (2006) Basal-like breast cancer and the BRCA1 phenotype Oncogene 25 5846–53 DOI: 10.1038/sj.onc.1209876 PMID: 16998499
52. Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K and Tlsty TD (2007) Abrogated response to cell stress identifies DCIS associated with subsequent tumour events and defines basal-like tumour Cancer Cell 12 479–91 DOI: 10.1016/j.ccr.2007.10.017 PMID: 17996651
53. Bohn OL, Fuertes-Camilo M, Navarro L, Saldivar J and Sanchez-Sosa S (2010) p16 INK4a expression in basal-like breast carcinoma Int J Clin Exp Pathol 3 600–07 PMID: 20661408 PMCID: 2907122
54. Subhawong AP, Subhawong T, Nassar H, Kouprina N, Begum S, Vang R, Westra WH and Argani P (2009) Most basal-like breast carcinomas demonstrate the same Rb−/ p16 immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically Am J Surg Pathol 33 163–75 DOI: 10.1097/PAS.0b013e31817f9790
55. Perou CM, Sørlie T, Eisen MB, Rijn M van de, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO and Botstein D (2000) Molecular portraits of human breast tumours Nature 406 747–52 DOI: 10.1038/35021093 PMID: 10963602
56. Ross DT and Perou CM (2001) A comparison of gene expression signatures from breast tumours and breast tissue derived cell lines Dis Markers 17 99–109 PMID: 11673656
57. Kotake Y, Cao R, Viatour P, Sage J, Zhang Yand Xiong Y(2007) pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumour suppressor gene Genes Dev 21 49–54 DOI: 10.1101/gad.1499407 PMID: 17210787 PMCID: 1759899