Unlocking artificial intelligence, machine learning and deep learning to combat therapeutic resistance in metastatic castration-resistant prostate cancer: a comprehensive review
Zainab Haider Ejaz1, Reyan Hussain Shaikh1, Alizeh Sonia Fatimi1 and Saqib Raza Khan2,3,a
1Aga Khan University Hospital, Aga Khan Medical College, Sindh, Karachi 748000, Pakistan
2Division of Medical Oncology, Department of Oncology, Schulich School of Medicine & Dentistry, Western University, London, ON N6A 3K7, Canada
3Verspeeten Family Cancer Centre, London Health Sciences Centre, London, ON N6A 5W9, Canada
ahttps://orcid.org/0000-0002-3742-534X
Abstract
Metastatic castration-resistant prostate cancer (mCRPC) remains a formidable clinical challenge despite advancements in therapy. This narrative review explores the role of artificial intelligence (AI), machine learning and deep learning in addressing therapeutic resistance in mCRPC. AI-driven approaches leverage integrated datasets encompassing genomics, proteomics and clinical parameters to uncover molecular mechanisms, predict treatment responses and identify biomarkers of resistance. These methodologies promise personalised treatment strategies tailored to individual patient profiles. However, data heterogeneity and regulatory considerations are challenges that hinder the translation of AI insights into clinical practice. By synthesising current literature, this review examines the progress, potential and limitations of AI applications in combating therapeutic resistance in mCRPC, highlighting implications for future research and clinical implementation.
Keywords: prostate cancer, castration resistance, metastasis, artificial intelligence, therapeutic resistance
Correspondence to: Saqib Raza Khan
Email: saqibrazakhan1994@gmail.com
Published: 29/07/2025
Received: 16/12/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Background
Castration-resistant prostate cancer (CRPC) is defined by the progression of prostate cancer, whether detected radiologically or biochemically, despite undergoing standard androgen deprivation therapy (ADT), where serum testosterone levels have decreased to castration levels (usually below 50 ng/dl or 1.7 nmol/l) [1]. Metastatic castration-resistant prostate cancer (mCRPC) is an aggressive form of cancer, prevalent among men and is associated with elevated morbidity and mortality [2]. With millions affected globally, mCRPC presents a formidable clinical challenge due to its aggressive nature and tendency to develop resistance to treatment. Despite advancements in oncological research and therapies, managing mCRPC remains difficult, as traditional approaches frequently fail to effectively halt disease progression or enhance patient outcomes [3].
The emergence of resistance mechanisms against frontline therapies, such as ADT and oral hormonal agents like enzalutamide and abiraterone underscore the urgent need for innovative strategies to address therapeutic resistance in mCRPC [4, 5]. The need for more effective interventions to combat this disease has spurred a growing interest in harnessing the power of artificial intelligence (AI), machine learning (ML) and deep learning (DL) to decipher its underlying complexities and devise personalised treatment regimens tailored to individual patient profiles [6-8].
In recent years, the convergence of AI, ML and DL technologies has propelled significant breakthroughs across various domains of healthcare, offering unprecedented opportunities for precision medicine, early disease detection and treatment optimisation [9]. Within the realm of mCRPC, the application of computational methodologies holds immense promise in elucidating the intricate molecular pathways driving therapeutic resistance and identifying novel targets for intervention. By analysing vast datasets encompassing genomics, proteomics and clinical parameters, AI-driven approaches can elucidate the molecular basis of treatment or disease progression or aid in the discovery of predictive biomarkers of the disease [10, 11].
This comprehensive review endeavors to explore the current landscape of AI-driven solutions aimed at overcoming therapeutic resistance in mCRPC.
Overview of artificial intelligence, machine learning and deep learning
In recent years, AI, ML and DL have garnered significant attention in oncology due to their potential to revolutionise research and clinical practice [12-14]. AI refers to the utilisation of a variety of techniques to enable computers to carry out simulated tasks requiring human intelligence [15].
ML, a subset of AI, involves the development of algorithms where computers learn the data and make predictions or decisions without explicit instructions [16, 17]. ML has been used in cancer research to analyse intricate data relationships and make predictions about cancer outcomes. Many studies have explored the utility of ML in cancer research and have highlighted its potential and limitations. Decision trees are a specific ML technique that has demonstrated broad applicability across various cancer types, significantly improving diagnostic accuracy [18-20]. These applications encompass various cancer types, including breast, gastric, thyroid, prostate and colorectal cancer. Additionally, other ML algorithms such as k-means, K-nearest neighbors [21], logistic regression [22], Naïve Bayes [23, 24], principal component analysis [25, 26], Random Forests [27, 28, 29] and eXtreme Gradient Boost [30] have also been employed in cancer research, contributing to improved diagnostic accuracy and prediction across various cancer types [31].
DL is a specialised form of ML where multilayered artificial neural networks are employed to extract complex features from vast datasets, enabling the creation of powerful models that are able to capture complicated patterns [32]. DL has emerged as a transformative technology in cancer research. It can learn intricate features from expansive datasets automatically, making it a powerful tool for analysing images and molecular profiling. Studies that have explored the application of DL have shown its remarkable potential in improving diagnosis, predicting disease progression and exploring new treatment options [33, 34]. Moreover, initiatives such as the introduction of expansive pathological image datasets, such as SNOW for breast cancer research, have significantly improved computational pathology by providing abundant data for training robust DL models [35]. Furthermore, the development of DL-based methods for the automatic identification of circulating tumour cells and cancer-associated fibroblasts has shown superior performance compared to conventional computer vision methods [35].
The widespread success of AI, ML and DL in improving diagnostics and treatment strategies across various cancer types underscores their potential utility in addressing the complexities of mCRPC. Evidence from studies in breast, gastric and colorectal cancer highlights the significant contributions of these technologies in enhancing diagnostic accuracy and predicting therapeutic responses. Utilising the capabilities of AI, ML and DL in mCRPC research and clinical practice could lead to personalised therapeutic interventions tailored to individual patient profiles, ultimately improving outcomes for those affected by this aggressive form of prostate cancer. Thus, the demonstrated efficacy of AI-driven approaches in other cancer types suggests a promising avenue for combating therapeutic resistance in mCRPC through interdisciplinary collaboration and innovative application of these methodologies.
Pathogenesis of metastatic castration-resistant prostate cancer
Patients diagnosed with localised prostate cancer usually undergo radical prostatectomy and/or radiotherapy, followed by ADT [36, 37]. Based on the cancer grade, a varying percentage of these patients progress to CRPC by one decade [38, 39]. CRPC, previously known as ‘hormone-refractory prostate cancer’ and ‘androgen-independent prostate cancer,’ still relies on hormone activity for androgen receptor (AR) activation, even though castration treatments like ADT were ineffective [40-42]. Consequently, the terms ‘hormone-refractory prostate cancer’ and ‘androgen-independent prostate cancer’ were replaced with ‘castration-resistant prostate cancer (CRPC)’ [43, 44].
It is widely recognised that patients with metastatic prostate cancer undergoing ADT tend to show disease progression after an average of 18–36 months [45]. The stem cell model of the prostate highlights that hormone resistance can arise through multiple pathways [46].
The normal prostate is maintained by a small number of androgen-independent stem cells that continually replenish themselves and produce androgen-sensitive cells [47]. When testosterone levels are within the physiological range, these androgen-sensitive cells differentiate into glandular epithelial cells until the cell population reaches equilibrium, balancing proliferation and cell death to prevent excessive growth [47]. As a result, the typical prostate consists predominantly of androgen-dependent glandular cells, a smaller number of androgen-sensitive basal cells and a limited population of androgen-independent basal stem cells [47].
Although the precise mechanisms are not entirely clear, several factors likely contribute to androgen independence, including genetic instability due to alterations in microenvironmental, impaired detoxification, activation of oncogenes and increased levels of androgen receptors [47].
Mechanisms of therapeutic resistance in mCRPC
The therapeutic landscape in mCRPC is predominantly limited to taxane-based chemotherapies, with emerging secondary regimens such as cabazitaxel and AR-targeted therapies like abiraterone, enzalutamide and apalutamide [43, 48]. However, resistance to these therapies poses a significant challenge, driving ongoing research into new targets and drug development strategies for mCRPC.
Resistance mechanisms in mCRPC are intricate and diverse. Understanding these mechanisms is crucial for devising effective therapeutic strategies. A primary mechanism involves the overexpression of efflux proteins such as P-glycoprotein and multidrug resistance proteins, which expel the chemotherapy drugs from tumour cells, decreasing their effectiveness [43, 49].
The AR signalling pathway is involved in mCRPC progression, and resistance mechanisms against AR-targeted medications like abiraterone and enzalutamide are well-documented. Mutations in the ligand-binding domain (LBD) of AR can alter its conformation, causing receptor antagonists to act as agonists, thereby promoting resistance [49]. Up to 64% of mCRPC patients exhibit AR overexpression or amplification, while AR ovlike AR-V7, lacking the LBD, remain constitutively active and contribute further to therapeutic resistance [50, 51]. AR overexpression could be due to overexpression of AR coregulators, increased stability of the receptor itself or gene amplification resulting in increased phosphorylation or acetylation of histones at AR enhancers [52, 53]. Additionally, glucocorticoid receptor (GR) upregulation has been implicated in enzalutamide resistance, as both AR and GR share a binding site on the chromosome, leading to the activation of AR-specific genes [54, 55].
Another critical resistance mechanism involves tumour cells evading destruction by inhibiting apoptosis and repairing DNA damage, processes regulated by non-coding RNAs that modulate gene expression [56] Epigenetic changes also play a key role in mCRPC, such as covalent modifications of histone proteins, which ultimately affect the normal processes associated with DNA, thereby playing a role in disease progression [57].
Neuroendocrine differentiation of tumour cells promotes lineage plasticity, whereby cells show increased adaptability to survival by adapting to their environment [58, 59]. This concept of plasticity has also been suggested as a possible mechanism of therapeutic resistance and disease progression [60].
Activation of PI3K and AKT in mCRPC offers alternative survival pathways for the tumour cells, promoting cell growth and multiplication [61, 62]. The prostate tumour microenvironment is another contributing factor, involving a complex interaction between tumour cells, stromal cells and various immune components [63]. This environment is highly immunosuppressive, driven by inflammatory signals and low oxygen levels, which create a protective barrier around cancer cells [63]. Immune-suppressive cells, such as regulatory T cells, M2 macrophages and myeloid-derived suppressor cells, dominate the microenvironment, impeding the anti-tumour activities of crucial immune cells like dendritic cells, natural killer cells, B cells and cytotoxic T cells [63]. This suppression ultimately leads to resistance against treatments.
Poly-ADP ribose polymerase (PARP) inhibitors may also be used in treating mCRPC, but resistance mechanisms including aberrations in genes of homologous recombination repair (HRR) such as BRCA1, BRCA2 and PALB2, pose significant challenges [64]. Efflux transporters like ABC transporters also reduce intracellular concentrations of PARP inhibitors, contributing to resistance [65].
Despite early success, resistance to 177 Lu-PSMA radioligand therapy in mCRPC arises from its low linear energy transfer, causing primarily single-strand DNA breaks. In addition, the effectiveness of this therapy depends on PSMA-mediated uptake of the radioligand; tumours with low or heterogeneous PSMA expression may not receive adequate radiation doses, leading to reduced therapeutic efficacy and prostate-specific antigen (PSA) response [66].
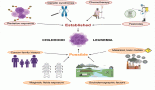
These examples, summarised in Figure 1, underscore the complexity of resistance mechanisms in mCRPC, highlighting the need for innovative approaches to overcome them.
Current standard of care in the treatment of metastatic castration-resistant prostate cancer
The National Comprehensive Cancer Network (NCCN) guidelines offer a multifaceted approach to managing mCRPC, advocating for the continuation of ADT alongside oral hormonal agents, chemotherapies such as docetaxel and oral targeted agents including abiraterone and enzalutamide, which are preferred choices if not previously administered [67].
Clinical decision-making in mCRPC hinges on various factors including prior treatments, patient preferences, symptomatology, adverse effects and the presence of visceral disease [67]. Regular monitoring via radiographic imaging (e.g., Computerised Tomography (CT) scans, bone imaging, prostate-specific membrane antigen positron emission tomography (PSMA PET) scans), clinical examinations and laboratory tests (e.g., PSA) are essential [67]. Therapy is typically continued until disease progression or intolerable side effects develop, as per NCCN guidelines [67].
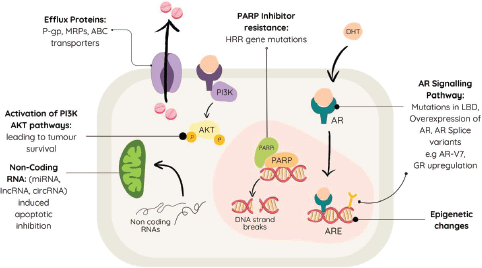
Figure 1. Various methods of therapeutic resistance in metastatic castration-resistant prostate cancer.
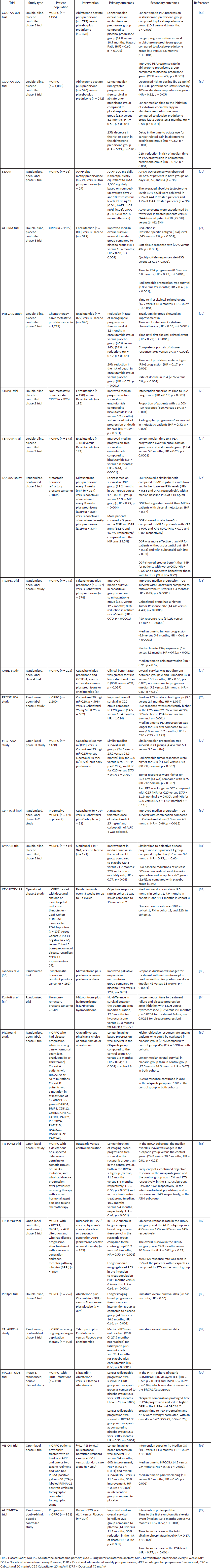
Numerous novel therapies have been tested in clinical trials, summarised in Table 1, offering potential avenues for mCRPC treatment. Despite their demonstrated efficacy, overall survival rates remain low, highlighting the ongoing necessity for extensive clinical trials to establish definitive outcomes and for the developing new drugs and therapies to combat this problem.
Table 1. Overview of novel therapies investigated in clinical trials for mCRPC
Applications of artificial intelligence, machine learning and deep learning in combating therapeutic resistance
AI holds immense potential to transform the landscape of mCRPC treatment by revolutionising various aspects of patient care. Figure 2 illustrates the diverse applications of AI in targeting mCRPC. AI algorithms can analyse patient data, including genomic profiles, imaging results and treatment histories, to predict individual responses to various treatment modalities. By identifying individual patient characteristics, clinicians can create customised treatment plans that optimise therapeutic benefits while reducing adverse effects.
Table 2 summarises the current literature on AI applications in the diagnosis and treatment of prostate cancer.
Pathomics
While prostate biopsy and Gleason scoring have historically served as the cornerstone for diagnosing localised prostate cancer, they are typically performed early in the disease course and often precede the development of mCRPC by several years [93]. Although prostate biopsy is not routinely required for the diagnosis of mCRPC, it retains a critical role in select clinical scenarios, particularly for molecular profiling and therapeutic stratification. Biopsies of metastatic sites are increasingly employed to enable mutational analysis and immunohistochemistry, identifying actionable genomic alterations such as BRCA1/2, ATM and MSI-H, which can guide personalised treatment decisions [94]. Moreover, tissue biopsy plays a role in ruling out neuroendocrine or small cell transformation, which warrants a distinct therapeutic approach [95]. Despite 58% of patients in one study being found to harbor theoretically actionable mutations, only a small subset received matched therapies, highlighting the disparity between molecular findings and clinical application [94]. Beyond traditional histopathology, AI has demonstrated considerable promise in enhancing pathology workflows. In a meta-analysis by Morozov et al [93], AI-assisted histological assessment across 8,000 prostate biopsies and 458 prostatectomy cases achieved diagnostic accuracies ranging from 83.7% to 98.3%. Similarly, the PANDA challenge led by Bulten et al [96], utilising over 10,000 digitised biopsies, showcased AI’s potential to improve the reproducibility and efficiency of Gleason grading.
Jung et al [97] validated DeepDx® Prostate (DeepDx), an AI-based diagnostic tool, using 593 whole-slide images (130 normal, 463 adenocarcinomas) against Gleason scores and grade groups assessed by three expert uropathologists as the reference standard [97]. DeepDx demonstrated comparable cancer detection accuracy to original pathology reports but showed higher concordance with reference grade groups and Gleason scores. In another study, Paige Prostate, a clinical-grade AI tool, was evaluated for its efficacy in aiding pathologists with identification, grading and quantifying prostate cancer in 105 prostate core needle biopsies (CNBs) [100]. Four pathologists initially diagnosed the CNBs independently, achieving a diagnostic accuracy of 95.0%.
While these advancements have primarily focused on localised disease, their integration into molecular pathology pipelines and application to metastatic lesion evaluation remain promising avenues for optimising precision oncology in mCRPC. In addition, early detection of prostate cancer allows for timely intervention, significantly impacting mCRPC prognosis. Detecting prostate cancer early increases the likelihood of successful localised treatment with surgery or radiation therapy, potentially preventing or delaying progression to advanced stages, including mCRPC. This timely intervention facilitates the early initiation of systemic therapies and enables more effective disease monitoring and management. Figure 3 summarises the benefits and potential challenges of incorporating AI into pathomics related to mCRPC diagnosis.
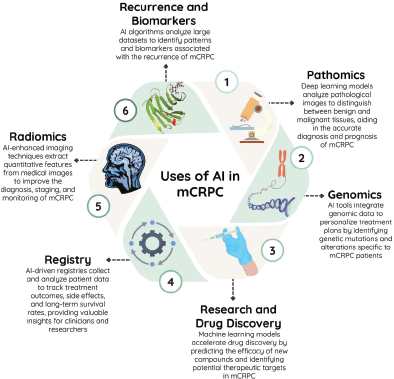
Figure 2. Different applications of artificial intelligence in the treatment of metastatic castration-resistant prostate cancer.
Table 2. Current literature available on AI applications in diagnosis, grading and treatment of prostate cancer.
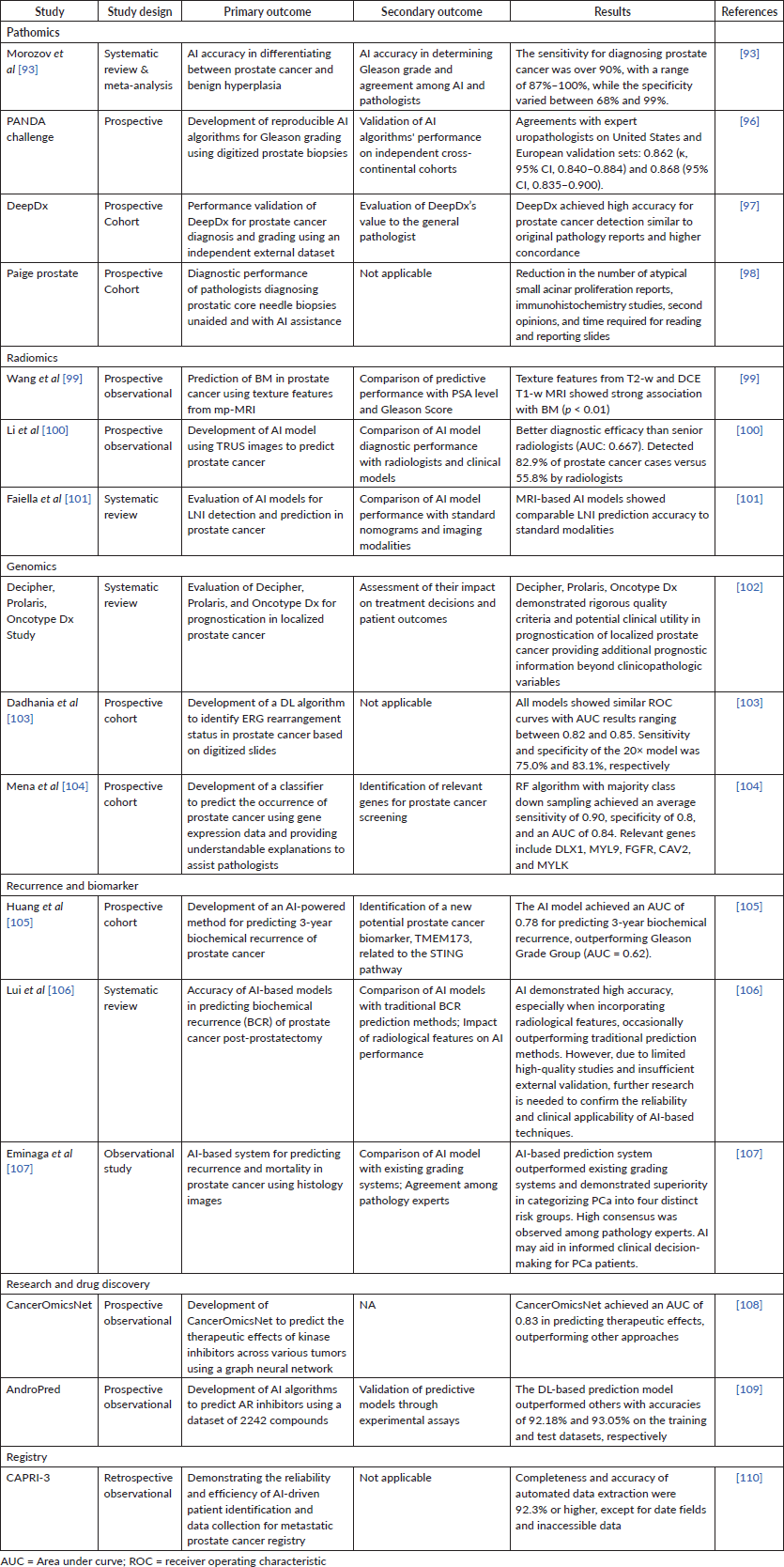
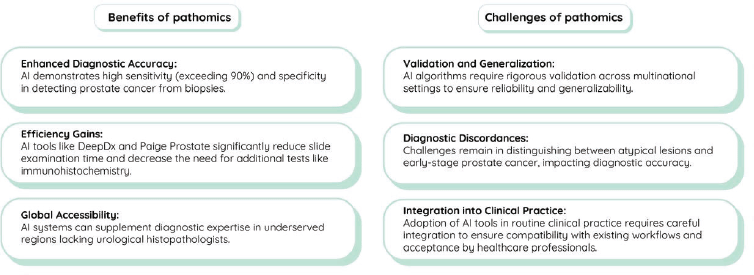
Figure 3. Benefits and challenges of pathomics in the diagnosis of metastatic castration-resistant prostate cancer.
Radiomics
The field of radiomics, which involves developing computational systems to extract and analyse quantitative features from medical images, holds promise for timely metastasis detection in mCRPC.
Wang et al [99] investigated the utility of texture features taken from multiparametric magnetic resonance imaging (mp-MRI) to predict bone metastases in prostate cancer patients [99]. By analysing 976 features from T2-weighted (T2-w) and dynamic contrast-enhanced T1-weighted (DCE T1-w) MRI scans of 176 patients, they found that combining information from both MRI sequences showed improved prognostic performance compared to using either sequence alone. Another study by Li et al [100] focused on developing an AI model using transrectal ultrasonography (TRUS) images to predict prostate cancer, comparing its diagnostic performance to radiologists and a clinical model [100]. Using 1,696 2-dimensional TRUS images from 142 patients, they trained a ResNet50 network with three classification models: original image (Whole), biopsy needle tract (Needle) and combined image (Feature Pyramid Networks (FPN)). The FPN model outperformed others.
Faiella et al [101] examined the role of AI models in detecting and predicting lymph node involvement (LNI) in prostate cancer patients. They evaluated 16 studies, where AI models predicted LNI as accurately as traditional nomograms, while CT and PET-CT based models showed strong diagnostic and prognostic capabilities.
Radiomics' ability to analyse texture features from medical imaging offers early detection potential for BM in mCRPC, aiding in treatment planning and prognosis assessment. Its quantitative approach complements traditional imaging modalities, potentially enhancing sensitivity and specificity, thereby advancing precision medicine in mCRPC management. Figure 4 outlines anticipated challenges associated with radiomics in mCRPC and proposes strategies to overcome them.
Genomics
Decipher, Oncotype DX Genomic Prostate Score and Prolaris are commercially available genomic tests, powered by ML, that guide prostate cancer management [102]. Decipher analyses gene expression to predict disease recurrence post-treatment. Oncotype DX Genomic Prostate Score assesses tumour aggressiveness to guide treatment decisions, especially post-surgery. Prolaris measures cell proliferation genes to predict disease aggressiveness and guide treatment strategies. These classifiers enable personalised treatment plans, sparing low-risk patients from unnecessary interventions while identifying high-risk cases needing more aggressive therapy. By integrating genomic data with clinical parameters, they enhance precision medicine, improving patient outcomes and quality of life. Clinicians can utilise these tools to tailor treatments, reducing overtreatment and minimising side effects. Their adoption signifies a shift towards data-driven decision-making in oncology, optimising patient care by harnessing the power of ML and genomics.
Dadhania et al [103] used digitised slides of hematoxylin and eosin staining and identified the rearrangement of TMPRSS2-ERG in prostate cancer using a DL algorithm. Image patches from slides from 392 cases were exported at various magnifications. A DL model based on MobileNetV2 architecture was trained separately for each magnification level. The area under the curve (AUC) of the receiver operating characteristic (ROC) curves ranged between 0.82 and 0.85. The sensitivity of the models was 75.0% and the specificity was 83.1% (20 × model). Mena et al [104] developed an ML-based classifier using 47 genes selected for their differential expression in prostate cancer, gene ontology and supporting literature. This classifier was trained using data available from 550 samples obtained from 'The Cancer Genome Atlas' and subsequently validated on four diverse external datasets comprising a total of 463 samples. Strong statistical significance was shown for the most successful strategy, which combined the Random Forest method with majority class downsampling. Across all datasets, their approach consistently produced an average sensitivity of 0.90, specificity of 0.80 and AUC of 0.84.
Genomics offers a promising opportunity for personalised treatment strategies. By identifying specific gene mutations or rearrangements, the most ideal drug therapy can be suggested and its efficacy can be predicted beforehand. For instance, genomic profiling can reveal mutations in DNA repair genes like BRCA1/2 or alterations in the AR signalling pathway, which can guide the use of targeted therapies such as PARP inhibitors for BRCA-mutated cancers or next-generation AR signalling inhibitors. Moreover, clinical trials can be enhanced by stratifying the participants on genomic data. By focusing on genomic-driven drug development, newer and more effective therapies can emerge for mCRPC.
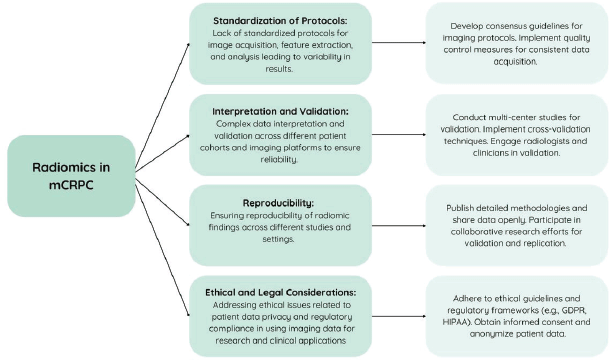
Figure 4. Challenges of incorporating radiomics in the treatment of metastatic castration-resistant prostate cancer and their solutions.
Recurrence prediction
Huang et al [105] presented a novel AI-powered method for predicting an early recurrence of prostate cancer post-prostatectomy and identifying cancer driver regions. Using deep convolutional neural networks, the study developed an AI model trained on whole slide images and patient data. The model extracted both visual and microscopic morphological features to find predictive regions of early recurrence (regions of interest [ROIs]). The model demonstrated promising results, with AI-derived morphometric scores effectively ranking regions of ROIs and showing strong prognostic value for 3-year biochemical recurrence (AUC = 0.78), significantly outperforming the traditional Gleason Grade Group, which had an AUC of 0.62.
Liu et al [106] in their systematic review evaluates the effectiveness of AI in predicting biochemical recurrence after prostate cancer surgery. AI algorithms, especially those using radiological features, demonstrated higher accuracy in predicting the recurrence compared to those based on pathological or clinicopathological data. In some cases, AI outperformed traditional prediction methods. However, the review also identified significant variability in AI performance due to differences in study designs, patient inclusion criteria and follow-up data.
Eminaga et al [107] developed an AI-based system to predict prostate cancer recurrence using histology images. Validated with multi-institutional datasets of 2,647 patients over 10 years, the system demonstrated superior predictive accuracy compared to existing grading systems. It categorised prostate cancer into four distinct risk groups and showed high consensus among experts, suggesting that AI can enhance recurrence prediction and improve clinical decision-making for patients.
These studies underscore the potential of AI to accurately predict the recurrence of prostate cancer.
Biomarker discovery
In addition, Huang et al [105] identified a potential new prostate cancer biomarker, TMEM173, associated with the STING pathway, through focused biomarker analysis of high-scored ROIs. This research introduced an innovative approach to identifying prostate cancer patients at risk for early recurrence and discovering novel biomarkers, utilising AI to analyse morphologic features from whole slide image data.
Research and drug discovery
CancerOmicsNet is a novel system based on AI that predicts how well the multitargeted kinase inhibitors will work against various types of cancer using a deep graph learning model [108]. Gagare et al [109] utilised ML and DL algorithms to train predictive models using molecular features, achieving high accuracy on both training and test datasets. It is important to further validate these models using experimental assays to establish their full reliability and utility.
Nevertheless, AI algorithms can analyse large-scale biomedical data, including genomic data and clinical trial results, to identify potential biomarkers, therapeutic targets and novel drug candidates for prostate cancer. This accelerates the process of drug development, allowing more effective therapies.
Registry management
Maintaining an efficient registry for metastatic prostate cancer is crucial for providing the latest data to clinicians. However, using electronic health records (EHRs) that are mainly present in the form of text, to manage patient data poses challenges. Recent advancements in AI technologies, such as named entity recognition and natural language processing (NLP) have enabled the acquisition of valuable information from large amounts of unstructured textual data (text mining) [110]. In the Netherlands, NLP-based text-mining software is widely used across hospitals to semiautomatically gather data from EHRs [110]. This registry enables clinicians and researchers to access updated data, analyse treatment trends, monitor disease progression and evaluate patient outcomes. Access to such data is critical for making informed treatment decisions, modifying strategies based on the latest outcomes and adapting to emerging trends in patient care.
Ongoing trials incorporating artificial intelligence to develop robust algorithms in the management of prostate cancer
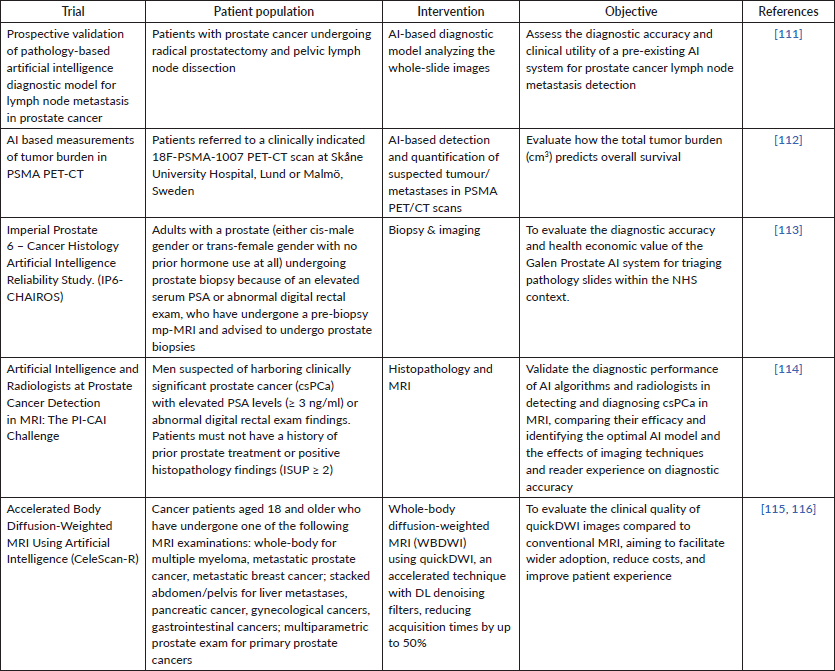
Currently, multiple trials are ongoing in oncology and AI (Table 3). By incorporating AI with diagnostic imaging, these trials hold immense potential for developing robust algorithms and models that can facilitate accurate and prompt diagnosis. Continuous follow-up of these trials can provide valuable insights into treatment responses and assist in predicting therapeutic outcomes based on AI's ability to analyse images. This integration of AI offers an opportunity to adjust therapeutic strategies, allowing for the most suitable treatment approach for the patient without compromising the quality of healthcare.
Table 3. Ongoing trials incorporating artificial intelligence to develop robust algorithms in the management of prostate cancer.
Studies involving artificial intelligence to reduce therapeutic resistance in CRPC
Several studies have explored the integration of AI to understand therapeutic resistance and the effects of various therapies on a cellular level in CRPC. For instance, Blatti et al [10] developed a tool called TraRe, which used RNA sequencing to determine the response of abiraterone in mCRPC in different transcriptional networks. TraRe pinpointed a specific transcriptional module linked to the immune response, enriched with rewired regulatory networks in abiraterone treatment responders versus non-responders. This module included key transcription factors such as CEBPE, GATA1, KLF1 and MYB, known to influence granulocyte differentiation, erythroid development and various hematopoietic and immune pathways. Additionally, TAL1 and NFE2 emerged as pivotal factors in gene expression regulation. Furthermore, TraRe identified transcription factors like SREBF2, SMAD7, SOX8 and SNAI2 driving other rewired transcriptional modules potentially implicated in cancer progression and resistance mechanisms. The analysis also highlighted the crucial role of three transcription factors (ELK3, MYB and MXD1) which functioned differently in patients who didn't respond well to treatment. These findings underscore the complexity of transcriptional rewiring in mCRPC and the potential utility of TraRe in formulating personalised treatment plans.
Xue et al [117] predicted dosages using an ML technique for mCRPC patients. The patients’ PET imaging data and clinical information were used in the ML model as inputs, while Hermes software was utilised to calculate the organ-level dosimetry. Using standard uptake values, the ML algorithms predicted the dosimetry at the organ level. This algorithm can potentially improve treatment for CRPC by personalising radiation plans to maximise tumour kill while minimising damage to healthy organs. Through the prediction of individual radiation tolerance, this method may enable more aggressive dosing without compromising safety thresholds.
AI models can also be used to predict treatment discontinuity due to adverse effects. An example of this would be the algorithm laid out by Deng et al [118] which integrated data from three cohorts from different clinical trials. Various ML models were tested, including linear regression, cox regression, logistic regression and nonlinear tree-based methods. Tree-based models performed better, with random forest (RF) showing the highest performance in two out of the three cohorts. This RF model was then trained to identify patterns that differentiate patients who discontinue treatment from those who continue, with certain data features, such as laboratory test results and medical history, having more influence on the model’s predictions. This model could help personalise treatment plans for CRPC, potentially reducing unwanted side effects and improving adherence and tolerance to treatment.
AI also has the ability to identify genomic mutations in mCRPC, which can then be used to identify different targets for drug development. One such study was conducted by Lin et al [119] which explored the next-generation sequencing of circulating cell-free DNA and used ML algorithms to identify specific genomic alterations to distinguish patients with castration-resistant from those with castration-sensitive prostate cancer (CSPC). This study revealed potential genomic alterations in patients with mCRPC, specifically in the PI3K, RTK, G1/S and MAPK signalling pathways, highlighting the need to potentially target these pathways for therapeutic purposes.
Challenges and limitations of successful implementation of artificial intelligence to combat therapeutic resistance in castration-resistant prostate cancer
The successful implementation of AI in healthcare, particularly in combating therapeutic resistance in CRPC, faces numerous challenges. Firstly, ML and DL algorithms rely on vast datasets, which often present problems since hospital data is considered confidential and not commonly shared between institutions [120, 121]. This challenge of data accessibility hinders the development of complex algorithms, especially because ML-based systems depend on continuous improvement through training with expanding datasets [122]. The large data required also necessitates the establishment of an AI-supporting platform that can hold large amounts of data for storage, processing and analysis [123]. This is critical to understanding and predicting therapeutic resistance patterns effectively. Unfortunately, establishing such platforms involves expensive hardware, software and skilled personnel, costs that are often too high for individual research teams to afford [123].
Heterogeneity in disease presentation and progression presents a special difficulty in the setting of prostate cancer, especially in mCRPC, which involves a variety of genetic subtypes (e.g., BRCA1/2-mutated, AR-V7 positive, neuroendocrine transformation). The generalisability of the model may be hampered by this variability. For example, when applied to mCRPC populations, where therapeutic resistance mechanisms differ dramatically from hormone-sensitive illness, AI models trained on localised prostate cancer datasets may perform poorly. Another limitation is data labeling and annotation complexity, especially in histopathology. While AI has demonstrated promise in tasks such as Gleason grading, training models to detect subtle features like neuroendocrine differentiation or intratumoural heterogeneity – hallmarks of advanced mCRPC – is far more complex, often requiring expert manual annotation and large sample diversity.
Another potential issue concerns patient consent and data privacy. A prominent example occurred in 2018 with the acquisition of DeepMind Health by Google. Their application, Streams, was created to manage patients with acute kidney injuries. However, the project faced criticism when it was revealed that the National Health Service (NHS) had shared sensitive data of more than 1 million patients with DeepMind servers without obtaining patient consent [124]. This underscores the critical issue of patient consent, as healthcare data can be easily breached given its sensitivity [125]. The emergence of AI adds to these complications, as individuals may inadvertently grant access to further data collection by mistaking AI systems for human interaction [126]. Thus, it is crucial for AI to play a safe and ethical role in healthcare by adhering to medical ethics and laws. Additionally, specific regulations governing the development and implementation of such technologies are warranted [127]. In mCRPC, patient consent and privacy are paramount given the confidentiality of cancer data.
AI algorithms are also not perfectly reliable, and uncorrected errors within AI systems can lead to negative outcomes that may have significant impacts [128]. This is exemplified by an AI application built to assess the risk of post-pneumonia complications, which malfunctioned and incorrectly recommended discharging asthmatic patients, posing a potential health risk [129]. In prostate cancer, incorrect interpretation of imaging or biopsy data could result in misclassification of disease stage or resistance profile, leading to suboptimal treatment selection. This raises a pertinent problem regarding accountability: who is responsible for the mishap when the AI system makes an error? With a lack of guidelines on the ethical use of AI/ML/DL algorithms in healthcare, there is ambiguity in their use in hospitals, leading to reluctance to implement these technologies [130].
As described by Laranjo et al [131] current research in the field of diagnosis using ML techniques often lacks consistency in reporting and fails to address the practical needs of end-users [131]. The current focus rests primarily on assessing technical performance using historical data, neglecting the crucial ‘last mile’ of clinical evaluations through randomised trials that assess real-world clinical use of AI [132, 133]. This creates a problem with a lack of replication of trials, limiting the evidential data and potentiating the risk of methodological error [134]. The capacity to independently validate research findings is the cornerstone of rigorous scientific investigation, which is accomplished by replication studies that effectively replicate the original experiment under controlled settings [131]. While there are currently limited replication studies in health informatics [135], the problem extends further with the fact that some replication studies are unable to produce the same results as the original study, creating problems regarding the clinical implementation of AI programs [136].
Another problem regarding AI technologies is the lack of transparency, classically described as the ‘black box problem’ [137, 138, 139]. DL algorithms often fail to provide justifications for their predictions, presenting legal difficulties as the system cannot justify potentially erroneous recommendations [140]. Lack of transparency also fails to establish patients’ trust in the system, posing problems regarding the practical implementation of these systems in clinics [141]. This is especially problematic in prostate cancer, where treatment decisions hinge on nuanced variables such as androgen receptor splice variants, genomic alterations and immunohistochemical profiles. Without clear justifications for AI outputs, clinicians may hesitate to trust these models, impeding clinical adoption.
Data heterogeneity presents another problem. Clinical data might differ greatly in terms of quality and presentation. This is best exemplified by surgical notes that are often varied and heterogeneous [123]. These differences may make it difficult for AI models trained on certain datasets to generalise to new data sources. This is compounded in prostate cancer and CRPC by variations in reporting standards across biopsy pathology, MRI protocols and molecular testing platforms, making generalisation across centers difficult. This highlights the need to maintain datasets containing cleaned data with a maximal signal-to-noise ratio to ensure robustness and reliability for the widespread application of AI techniques [123].
Another significant reason for the reluctance to adopt AI is the belief that AI will replace the human workforce [121]. This is a flawed perspective in medicine as it is a dynamic field with unpredictable circumstances that often require intuition and innate abilities to deal with various situations [142]. AI should be regarded as a valuable tool to support and strengthen clinical judgment, not as a substitute, helping oncologists tailor personalised treatment plans for mCRPC effectively.
While these problems exist universally, the implementation of AI technologies faces additional challenges in low- and middle-income countries (LMICs). In these regions, populations often lack access to basic healthcare due to financial constraints and poor infrastructure. With a significant proportion of the population residing in rural areas, the quality of healthcare provided is often mediocre to low. This is further augmented by the lack of well-developed roads for timely access to healthcare facilities. Additionally, these areas often lack access to internet facilities. While there are pertinent solutions, such as building infrastructure and installing internet devices, the costs required to achieve this impose an immense economic burden. This highlights how LMICs are bound by limited resources and finances, creating problems in the adoption of AI systems in these areas.
Training professionals, improving healthcare access and devising innovative solutions to provide AI-based systems in LMICs is essential and requires multidisciplinary, dedicated efforts. Specifically, for mCRPC, training healthcare providers on AI tools and methods to combat therapeutic resistance can significantly enhance treatment efficacy and patient outcomes in these resource-constrained settings.
Conclusion
The review article sheds light on the emergence of AI algorithms in prostate cancer. The successful examples of AI systems in various disciplines demonstrate how these systems can revolutionise the field in terms of the speed and accuracy of diagnosis, potentially allowing prompt intervention to delay or prevent the development of mCRPC. While treatment options for mCRPC are limited, AI offers immense potential to discover novel agents and predict the therapeutic effects of drugs. AI can be leveraged not only by physicians in their routine clinical practice to decide the best possible therapeutic approach for the patient but also provides numerous opportunities to integrate genomic data for personalised treatment. With its potential to be utilised across multiple disciplines, AI holds a promising role in combating therapeutic resistance in mCRPC.
List of abbreviations
AI, Artificial intelligence; AR, Androgen receptor; cfDNA, circulating cell-free DNA; CRPC, Castration-resistant prostate cancer; DL, Deep learning; LMICs, Low middle income countries; ML, Machine learning; NGS, Next-generation sequencing; NHS, National Health Service.
Acknowledgments
Not applicable.
Conflicts of interest
The authors declare no competing interests.
Funding
The author(s) received no financial support for the research, authorship and/or publication of this article.
Declarations: human and animal rights and informed consent
No human or animal studies were carried out by the authors for this article.
Ethical considerations
Ethical approval was not required for this study as it is a review paper.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Author contributions
ZHE, RHS and ASF contributed to the literature search and writing of the original draft, review and editing.
SRK contributed to the conceptualisation of the review and supervision of the writing process, including critically revising the work.
References
1.Zarour L and Alumkal J (2010) Emerging therapies in castrate-resistant prostate cancer Curr Urol Rep 11(3) 104 https://doi.org/10.1007/s11934-010-0104-x
2.Siegel RL, Miller KD, and Jemal A (2018) Cancer statistics, 2018 CA Cancer J Clin 68(1) 442 https://doi.org/10.3322/caac.21442
3.Huang X, Chau CH, and Figg WD (2012) Challenges to improved therapeutics for metastatic castrate resistant prostate cancer: from recent successes and failures J Hematol Oncol 5 35 https://doi.org/10.1186/1756-8722-5-35 PMID: 22747660 PMCID: 3425086
4.Watson PA, Arora VK, and Sawyers CL (2015) Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer Nat Rev Cancer 15(12) 701–711 https://doi.org/10.1038/nrc4016 PMID: 26563462 PMCID: 4771416
5.Verma S, Prajapati KS, and Kushwaha PP, et al (2020) Resistance to second generation antiandrogens in prostate cancer: pathways and mechanisms Cancer Drug Resist 3(4) 45 https://doi.org/10.20517/cdr.2020.45
6.Tapper W, Carneiro G, and Mikropoulos C, et al (2024) The application of radiomics and AI to molecular imaging for prostate cancer J Pers Med 14(3) 287 https://doi.org/10.3390/jpm14030287 PMID: 38541029 PMCID: 10971024
7.Satturwar S and Parwani AV (2024) Artificial intelligence-enabled prostate cancer diagnosis and prognosis: current state and future implications Adv Anat Pathol 31(2) 425 https://doi.org/10.1097/PAP.0000000000000425
8.Baydoun A, Jia AY, and Zaorsky NG, et al (2024) Artificial intelligence applications in prostate cancer Prostate Cancer Prostatic Dis 27(1) 37–45 https://doi.org/10.1038/s41391-023-00684-0
9.Bajwa J, Munir U, and Nori A, et al (2021) Artificial intelligence in healthcare: transforming the practice of medicine Future Healthc J 8(2) e188–e194 https://doi.org/10.7861/fhj.2021-0095 PMID: 34286183 PMCID: 8285156
10.Blatti C, de la Fuente J, and Gao H, et al (2023) Bayesian machine learning enables identification of transcriptional network disruptions associated with drug-resistant prostate cancer Cancer Res 83(8) 1361–1380 https://doi.org/10.1158/0008-5472.CAN-22-1910 PMID: 36779846 PMCID: 10102853
11.Zhang X, Nakajima K, and Mizokami A, et al (2024) Flare phenomenon visualized by 99mTc-bone scintigraphy has prognostic value for patients with metastatic castration-resistant prostate cancer Ann Nucl Med 38(6) 428–440 https://doi.org/10.1007/s12149-024-01914-8 PMID: 38478154 PMCID: 11108890
12.Musa IH, Afolabi LO, and Zamit I, et al (2022) Artificial intelligence and machine learning in cancer research: a systematic and thematic analysis of the top 100 cited articles indexed in Scopus database Cancer Control 29 5946 https://doi.org/10.1177/10732748221095946
13.Burke HB, Goodman PH, and Rosen DB, et al (1997) Artificial neural networks improve the accuracy of cancer survival prediction Cancer 79(4) 857–862 https://doi.org/10.1002/(SICI)1097-0142(19970215)79:4<857::AID-CNCR24>3.0.CO;2-Y PMID: 9024725
14.Zugazagoitia J, Guedes C, and Ponce S, et al (2016) Current challenges in cancer treatment Clin Therap 38(7) 26 https://doi.org/10.1016/j.clinthera.2016.03.026
15.Hamet P and Tremblay J (2017) Artificial intelligence in medicine Metabolism 69 11 https://doi.org/10.1016/j.metabol.2017.01.011
16.Mendonça MOK, Netto SL, and Diniz PSR, et al (2024) Machine learning: review and trends Signal Process Mach Learn Theory 2024 869–959 https://doi.org/10.1016/B978-0-32-391772-8.00019-3
17.Wazid M, Das AK, and Chamola V, et al (2022) Uniting cyber security and machine learning: advantages, challenges and future research ICT Express 8(3) 313–321 https://doi.org/10.1016/j.icte.2022.04.007
18.Alharbi F and Vakanski A (2023) Machine learning methods for cancer classification using gene expression data: a review Bioengineering 10(2) 173 https://doi.org/10.3390/bioengineering10020173 PMID: 36829667 PMCID: 9952758
19.Swanson K, Wu E, and Zhang A, et al (2023) From patterns to patients: advances in clinical machine learning for cancer diagnosis, prognosis, and treatment Cell 186(8) 35 https://doi.org/10.1016/j.cell.2023.01.035
20.Bhatt M and Shende P (2023) Advancement in machine learning: a strategic lookout from cancer identification to treatment Arch Comput Methods Eng 30(4) 1–16 https://doi.org/10.1007/s11831-023-09886-0
21.Song C and Li X (2022) Cost‐Sensitive KNN algorithm for cancer prediction based on entropy analysis Entropy 24(2) 253 https://doi.org/10.3390/e24020253
22.Song X, Liang K, and Li J (2023) WGRLR: a weighted group regularized logistic regression for cancer diagnosis and gene selection IEEE/ACM Trans Comput Biol Bioinform 20(2) 1563–1573 https://doi.org/10.1109/TCBB.2022.3203167
23.Xiong Y, Ye M, and Wu C (2021) Cancer classification with a cost-sensitive naive bayes stacking ensemble Comput Math Methods Med 2021 5556992 https://doi.org/10.1155/2021/5556992 PMID: 33986823 PMCID: 8093037
24.Wood A, Shpilrain V, and Najarian K, et al (2019) Private naive bayes classification of personal biomedical data: application in cancer data analysis Comput Biol Med 105 18 https://doi.org/10.1016/j.compbiomed.2018.11.018
25.Park HA, Edelmann D, and Canzian F, et al (2022) Predictive polygenic score for outcome after first-line oxaliplatin-based chemotherapy in colorectal cancer patients using supervised principal component analysis Cancer Epidemiol Biomark Prev 31(11) 2087–2091 https://doi.org/10.1158/1055-9965.EPI-22-0320
26.Dweekat OY and Lam SS (2022) Cervical cancer diagnosis using an integrated system of principal component analysis, genetic algorithm, and multilayer perceptron Healthcare 10(10) 2002 https://doi.org/10.3390/healthcare10102002 PMID: 36292449 PMCID: 9601935
27.Gu T, Han Y, and Duan R (2023) A transfer learning approach based on random forest with application to breast cancer prediction in underrepresented populations Pac Sympos Biocomput 2022 186–197 https://doi.org/10.1142/9789811270611_0018
28.Zhang H, Chi M, and Su D, et al (2023) A random forest-based metabolic risk model to assess the prognosis and metabolism-related drug targets in ovarian cancer Comput Biol Med 153 106432 https://doi.org/10.1016/j.compbiomed.2022.106432 PMID: 36608460
29.Zhang J, Yang X, and Chen J, et al (2023) Construction of a diagnostic classifier for cervical intraepithelial neoplasia and cervical cancer based on XGBoost feature selection and random forest model J Obstetr Gynaecol Res 49(1) 296–303 https://doi.org/10.1111/jog.15458
30.Li B, Zhang F, and Niu Q, et al (2023) A molecular classification of gastric cancer associated with distinct clinical outcomes and validated by an XGBoost-based prediction model Mol Therapy Nucl Acids 31 14 https://doi.org/10.1016/j.omtn.2022.12.014
31.Zadeh Shirazi A, Tofighi M, and Gharavi A, et al (2024) The application of artificial intelligence to cancer research: a comprehensive guide Technol Cancer Res Treat 23 15330338241250324 https://doi.org/10.1177/15330338241250324 PMID: 38775067 PMCID: 11113055
32.Bonetto R, Latzko V (2020) Machine learning Computing in communication networks: from theory to practice (Cambridge:Academic Press) https://doi.org/10.1016/B978-0-12-820488-7.00021-9 pp 135–67
33.Maurya S, Tiwari S, and Mothukuri MC, et al (2023) A review on recent developments in cancer detection using machine learning and deep learning models Biomed Signal Process Control 80 104398 https://doi.org/10.1016/j.bspc.2022.104398
34.Nasser M and Yusof UK (2023) Deep learning based methods for breast cancer diagnosis: a systematic review and future direction Diagnostics 13(1) 161 https://doi.org/10.3390/diagnostics13010161 PMID: 36611453 PMCID: 9818155
35.Ding K, Zhou M, and Wang H, et al (2023) A large-scale synthetic pathological dataset for deep learning-enabled segmentation of breast cancer Sci Data 10(1) 231 https://doi.org/10.1038/s41597-023-02125-y PMID: 37085533 PMCID: 10121551
36.Mottet N, Bellmunt J, and Bolla M, et al (2017) EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent Eur Urol 71(4) 3 https://doi.org/10.1016/j.eururo.2016.08.003
37.Heidenreich A, Aus G, and Bolla M, et al (2008) EAU guidelines on prostate cancer Eur Urol 53(1) 68–80 https://doi.org/10.1016/j.eururo.2007.09.002
38.Heidenreich A, Bastian PJ, and Bellmunt J, et al (2014) EAU guidelines on prostate cancer. Part II: treatment of advanced, relapsing, and castration-resistant prostate cancer Eur Urol 65(2) 467–479 https://doi.org/10.1016/j.eururo.2013.11.002
39.Cornford P, Bellmunt J, and Bolla M, et al (2017) EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: treatment of relapsing, metastatic, and castration-resistant prostate cancer Eur Urol 71(4) 630–642 https://doi.org/10.1016/j.eururo.2016.08.002
40.Ahmad K (2004) New progress in treatment of hormone-refractory prostate cancer Lancet Oncol 5(12) 708–715 https://doi.org/10.1016/S1470-2045(04)01641-9
41.Petrylak DP (2005) Future directions in the treatment of androgen-independent prostate cancer Urology 65 8–12 https://doi.org/10.1016/j.urology.2005.04.020 PMID: 15939077
42.Montgomery RB, Mostaghel EA, and Vessella R, et al (2008) Maintenance of intratumoral androgens in metastatic prostate cancer: A mechanism for castration-resistant tumor growth Cancer Res 68(11) 4447–4454 https://doi.org/10.1158/0008-5472.CAN-08-0249 PMID: 18519708 PMCID: 2536685
43.Cai M, Song XL, and Li XA, et al (2023) Current therapy and drug resistance in metastatic castration-resistant prostate cancer Drug Resist Updates 68 100962 https://doi.org/10.1016/j.drup.2023.100962
44.He L, Fang H, and Chen C, et al (2020) Metastatic castration-resistant prostate cancer: academic insights and perspectives through bibliometric analysis Medicine 99(15) e19760 https://doi.org/10.1097/MD.0000000000019760
45.Newling DWW, Denis L, and Vermeylen K (1993) Orchiectomy versus goserelin and flutamide in the treatment of newly diagnosed metastatic prostate cancer. Analysis of the criteria of evaluation used in the european organization for research and treatment of cancer–genitourinary group study 30853 Cancer 72(12) 3793–3798 https://doi.org/10.1002/1097-0142(19931215)72:12+<3793::AID-CNCR2820721706>3.0.CO;2-U PMID: 8252492
46.Isaacs JT (1999) The biology of hormone refractory prostate cancer: Why does it develop? Urol Clin North Am 26(2) 263–273 https://doi.org/10.1016/S0094-0143(05)70066-5
47.Heidenreich A (2003) Therapeutic options in hormone refractory prostate cancer Prostate Cancer 2003 249–267 https://doi.org/10.1007/978-3-642-56321-8_27
48.Conteduca V, Mosca A, and Brighi N, et al (2021) New prognostic biomarkers in metastatic castration-resistant prostate cancer Cells 10(1) 10193 https://doi.org/10.3390/cells10010193
49.Taplin ME, Bubley GJ, and Shuster TD, et al (1995) Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer N Engl J Med 332(21) 1393–1398 https://doi.org/10.1056/NEJM199505253322101 PMID: 7723794
50.Annala M, Vandekerkhove G, and Khalaf D, et al (2018) Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer Cancer Discov 8(4) 937 https://doi.org/10.1158/2159-8290.CD-17-0937
51.Kallio HML, Hieta R, and Latonen L, et al (2018) Constitutively active androgen receptor splice variants AR-V3, AR-V7 and AR-V9 are co-expressed in castration-resistant prostate cancer metastases Br J Cancer 119(3) 347–356 https://doi.org/10.1038/s41416-018-0172-0
52.Lakshmana G and Baniahmad A (2019) Interference with the androgen receptor protein stability in therapy-resistant prostate cancer Int J Cancer 144(8) 1775–1779 https://doi.org/10.1002/ijc.31818
53.Kim EH, Cao D, and Mahajan NP, et al (2020) ACK1–AR and AR–HOXB13 signaling axes: epigenetic regulation of lethal prostate cancers NAR Cancer 2(3) zcaa018 https://doi.org/10.1093/narcan/zcaa018 PMID: 32885168 PMCID: 7454006
54.Kach J, Conzen SD, and Szmulewitz RZ (2015) Targeting the glucocorticoid receptor in breast and prostate cancers Sci Transl Med 7(305) aac7531 https://doi.org/10.1126/scitranslmed.aac7531
55.Arora VK, Schenkein E, and Murali R, et al (2013) XGlucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade Cell 155(6) 1309–1322 https://doi.org/10.1016/j.cell.2013.11.012
56.Lipovich L, Johnson R, and Lin CY (2010) MacroRNA underdogs in a microRNA world: Evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA Biochim Biophys Acta 1799(9) 597–615 https://doi.org/10.1016/j.bbagrm.2010.10.001 PMID: 20951849
57.Conteduca V, Hess J, and Yamada Y, et al (2021) Epigenetics in prostate cancer: Clinical implications Transl Androl Urol 10(7) 3104–3116 https://doi.org/10.21037/tau-20-1339 PMID: 34430414 PMCID: 8350251
58.Nijhout HF (2003) Development and evolution of adaptive polyphenisms Evol Dev 5 9–18 https://doi.org/10.1046/j.1525-142X.2003.03003.x
59.Zou M, Toivanen R, and Mitrofanova A, et al (2017) Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer Cancer Discov 7(7) 736–749 https://doi.org/10.1158/2159-8290.CD-16-1174 PMID: 28411207 PMCID: 5501744
60.Meacham CE and Morrison SJ (2013) Tumour heterogeneity and cancer cell plasticity Nature 501(7467) 12624 https://doi.org/10.1038/nature12624
61.Robinson D, van Allen EM, and Wu YM, et al (2015) Integrative clinical genomics of advanced prostate cancer Cell 161(5) 1215–1228 https://doi.org/10.1016/j.cell.2015.05.001 PMID: 26000489 PMCID: 4484602
62.Grasso CS, Wu YM, and Robinson DR, et al (2012) The mutational landscape of lethal castration-resistant prostate cancer Nature 487(7406) 11125 https://doi.org/10.1038/nature11125
63.Zhang Y, Campbell BK, and Stylli SS, et al (2022) The prostate cancer immune microenvironment, biomarkers and therapeutic intervention Uro 2(2) 74–92 https://doi.org/10.3390/uro2020010
64.Goodall J, Mateo J, and Yuan W, et al (2017) Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition Cancer Discov 7(9) 1006–1017 https://doi.org/10.1158/2159-8290.CD-17-0261 PMID: 28450425 PMCID: 6143169
65.Rottenberg S, Jaspers JE, and Kersbergen A, et al (2008) High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs Proc Natl Acad Sci U S A 105(44) 2105 https://doi.org/10.1073/pnas.0806092105
66.Arbuznikova D, Eder M, and Grosu AL, et al (2023) Towards improving the efficacy of PSMA-targeting radionuclide therapy for late-stage prostate cancer—combination strategies Curr Oncol Rep 25(11) 6 https://doi.org/10.1007/s11912-023-01458-6
67.Schaeffer EM, Srinivas S, and Adra N, et al (2024) NCCN guidelines® insights: prostate cancer, version 3.2024 J Natl Compr Canc Netw 22(3) 140–150 https://doi.org/10.6004/jnccn.2024.0019 PMID: 38626801
68.de Bono JS, Logothetis CJ, and Molina A, et al (2011) Abiraterone and increased survival in metastatic prostate cancer N Engl J Med 364(21) 618 https://doi.org/10.1056/NEJMoa1014618
69.Ryan CJ, Smith MR, and de Bono JS, et al (2013) Abiraterone in metastatic prostate cancer without previous chemotherapy N Engl J Med 368(2) 138–148 https://doi.org/10.1056/NEJMoa1209096 PMCID: 3683570
70.Stein CA, Levin R, and Given R, et al (2018) Randomized phase 2 therapeutic equivalence study of abiraterone acetate fine particle formulation vs. originator abiraterone acetate in patients with metastatic castration-resistant prostate cancer: the STAAR study Urol Oncol 36(2) 81.e9–81.e16 https://doi.org/10.1016/j.urolonc.2017.10.018
71.Scher HI, Fizazi K, and Saad F, et al (2012) Increased survival with enzalutamide in prostate cancer after chemotherapy N Engl J Med 367(13) 1187–1197 https://doi.org/10.1056/NEJMoa1207506 PMID: 22894553
72.Beer TM, Armstrong AJ, and Rathkopf DE, et al (2014) Enzalutamide in metastatic prostate cancer before chemotherapy N Engl J Med 371(5) 424–433 https://doi.org/10.1056/NEJMoa1405095 PMID: 24881730 PMCID: 4418931
73.Penson DF, Armstrong AJ, and Concepcion R, et al (2016) Enzalutamide versus bicalutamide in castration-resistant prostate cancer: The STRIVE trial J Clin Oncol 34(18) 2098–2106 https://doi.org/10.1200/JCO.2015.64.9285 PMID: 26811535
74.Shore ND, Chowdhury S, and Villers A, et al (2016) Efficacy and safety of enzalutamide versus bicalutamide for patients with metastatic prostate cancer (TERRAIN): A randomised, double-blind, phase 2 study Lancet Oncol 17(2) 153–163 https://doi.org/10.1016/S1470-2045(15)00518-5 PMID: 26774508
75.Berthold DR, Pond GR, and Soban F, et al (2008) Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer: Updated survival in the TAX 327 study J Clin Oncol 26(2) 242–245 https://doi.org/10.1200/JCO.2007.12.4008 PMID: 18182665
76.de Bono JS, Oudard S, and Ozguroglu M, et al (2010) Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: A randomised open-label trial The Lancet 376(9747) 1147–1154 https://doi.org/10.1016/S0140-6736(10)61389-X
77.Annala M, Fu S, and Bacon JVW, et al (2021) Cabazitaxel versus abiraterone or enzalutamide in poor prognosis metastatic castration-resistant prostate cancer: a multicentre, randomised, open-label, phase II trial Ann Oncol 32(7) 205 https://doi.org/10.1016/j.annonc.2021.03.205
78.Eisenberger M, Hardy-Bessard AC, and Kim CS, et al (2017) Phase III study comparing a reduced dose of cabazitaxel (20 mg/m2) and the currently approved dose (25 mg/m2) in postdocetaxel patients with metastatic castration-resistant prostate cancer - PROSELICA J Clin Oncol 35(28) 1076 https://doi.org/10.1200/JCO.2016.72.1076
79.Oudard S, Fizazi K, and Sengeløv L, et al (2017) Cabazitaxel versus docetaxel as first-line therapy for patients with metastatic castration-resistant prostate cancer: a randomized phase III trial - FIRSTANA J Clin Oncol 35(28) 1068 https://doi.org/10.1200/JCO.2016.72.1068
80.Corn PG, Heath EI, and Zurita A, et al (2019) Cabazitaxel plus carboplatin for the treatment of men with metastatic castration-resistant prostate cancers: a randomised, open-label, phase 1–2 trial Lancet Oncol 20(10) 1432–1443 https://doi.org/10.1016/S1470-2045(19)30408-5 PMID: 31515154 PMCID: 6858999
81.Kantoff PW, Higano CS, and Shore ND, et al (2010) Sipuleucel-T immunotherapy for castration-resistant prostate cancer N Engl J Med 363(5) 1294 https://doi.org/10.1056/NEJMoa1001294
82.Antonarakis ES, Piulats JM, and Gross-Goupil M, et al (2020) Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: Multicohort, open-label phase II KEYNOTE-199 study J Clin Oncol 38 1638 https://doi.org/10.1200/JCO.19.01638
83.Tannock IF, Osoba D, and Stockler MR, et al (1996) Chemotherapy with mitoxantrone plus prednisone or prednisone alone for symptomatic hormone-resistant prostate cancer: a Canadian randomized trial with palliative end points J Clin Oncol 14(6) 1756–1764 https://doi.org/10.1200/JCO.1996.14.6.1756 PMID: 8656243
84.Kantoff PW, Halabi S, and Conaway M, et al (1999) Hydrocortisone with or without mitoxantrone in men with hormone- refractory prostate cancer: results of the cancer and leukemia group B 9182 study J Clin Oncol 17(8) 2506 https://doi.org/10.1200/JCO.1999.17.8.2506 PMID: 10561316
85.de Bono J, Mateo J, and Fizazi K, et al (2020) Olaparib for metastatic castration-resistant prostate cancer N Engl J Med 382(22) 1440 https://doi.org/10.1056/NEJMoa1911440
86.Fizazi K, Piulats JM, and Reaume MN, et al (2023) Rucaparib or physician’s choice in metastatic prostate cancer N Engl J Med 388(8) 719–732 https://doi.org/10.1056/NEJMoa2214676 PMID: 36795891 PMCID: 10064172
87.Shah AS (2024) Is rucaparib the definite direction for metastatic prostate cancer? – TRITON3 results decoded Indian J Urol 40(1) 70–71 https://doi.org/10.4103/iju.iju_373_23 PMID: 38314070 PMCID: 10836449
88.Clarke NW, Armstrong AJ, and Thiery-Vuillemin A, et al (2022) Abiraterone and olaparib for metastatic castration-resistant prostate cancer NEJM Evid 1(9) EVIDoa2200043 https://doi.org/10.1056/EVIDoa2200043 PMID: 38319800
89.Agarwal N, Azad AA, and Carles J, et al (2023) Talazoparib plus enzalutamide in men with first-line metastatic castration-resistant prostate cancer (TALAPRO-2): a randomised, placebo-controlled, phase 3 trial Lancet 402 10398 https://doi.org/10.1016/S0140-6736(23)01055-3
90.Chi KN, Rathkopf D, and Smith MR, et al (2023) Niraparib and abiraterone acetate for metastatic castration-resistant prostate cancer J Clin Oncol 41(18) 1649 https://doi.org/10.1200/JCO.22.01649
91.Chi KN, Yip SM, and Bauman G, et al (2024) 177Lu-PSMA-617 in metastatic castration-resistant prostate cancer: a review of the evidence and implications for canadian clinical practice Curr Oncol 31(3) 106 https://doi.org/10.3390/curroncol31030106
92.Parker C, Nilsson S, and Heinrich D, et al (2013) Alpha emitter radium-223 and survival in metastatic prostate cancer N Engl J Med 369(3) 213–223 https://doi.org/10.1056/NEJMoa1213755 PMID: 23863050
93.Morozov A, Taratkin M, and Bazarkin A, et al (2023) A systematic review and meta-analysis of artificial intelligence diagnostic accuracy in prostate cancer histology identification and grading Prostate Cancer Prost Dis 26(4) 681–692 https://doi.org/10.1038/s41391-023-00673-3
94.Tao DL, Bailey S, and Beer TM, et al (2017) Molecular testing in patients with castration-resistant prostate cancer and its impact on clinical decision making JCO Precis Oncol 1 67 https://doi.org/10.1200/po.16.00067
95.Nadal R, Schweizer M, and Kryvenko ON, et al (2014) Small cell carcinoma of the prostate Nat Rev Urol 11(4) 213–219 https://doi.org/10.1038/nrurol.2014.21 PMID: 24535589 PMCID: 4339095
96.Bulten W, Kartasalo K, and Chen PHC, et al (2022) Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge Nat Med 28(1) 154–163 https://doi.org/10.1038/s41591-021-01620-2 PMID: 35027755 PMCID: 8799467
97.Jung M, Jin MS, and Kim C, et al (2022) Artificial intelligence system shows performance at the level of uropathologists for the detection and grading of prostate cancer in core needle biopsy: an independent external validation study Mod Pathol 35(10) 1449–1457 https://doi.org/10.1038/s41379-022-01077-9 PMID: 35487950
98.Eloy C, Marques A, and Pinto J, et al (2023) Artificial intelligence–assisted cancer diagnosis improves the efficiency of pathologists in prostatic biopsies Virchows Archiv 482(3) 1–10 https://doi.org/10.1007/s00428-023-03518-5
99.Wang Y, Yu B, and Zhong F, et al (2019) MRI-based texture analysis of the primary tumor for pre-treatment prediction of bone metastases in prostate cancer Magn Reson Imag 60 7 https://doi.org/10.1016/j.mri.2019.03.007
100.Li S, Ye X, and Tian H, et al (2024) An artificial intelligence model based on transrectal ultrasound images of biopsy needle tract tissues to differentiate prostate cancer Postgrad Med J 100(1182) 228–236 https://doi.org/10.1093/postmj/qgad127
101.Faiella E, Vaccarino F, and Ragone R, et al (2023) Can machine learning models detect and predict lymph node involvement in prostate cancer? a comprehensive systematic review J Clin Med 12(22) 7032 https://doi.org/10.3390/jcm12227032 PMID: 38002646 PMCID: 10672480
102.Ingrosso G, Alì E, and Marani S, et al (2022) Prognostic genomic tissue-based biomarkers in the treatment of localized prostate cancer J Pers Med 12(1) 10065 https://doi.org/10.3390/jpm12010065
103.Dadhania V, Gonzalez D, and Yousif M, et al (2022) Leveraging artificial intelligence to predict ERG gene fusion status in prostate cancer BMC Cancer 22(1) 494 https://doi.org/10.1186/s12885-022-09559-4 PMID: 35513774 PMCID: 9069768
104.Ramírez-Mena A, Andrés-León E, and Alvarez-Cubero MJ, et al (2023) Explainable artificial intelligence to predict and identify prostate cancer tissue by gene expression Comput Methods Prog Biomed 240 107719 https://doi.org/10.1016/j.cmpb.2023.107719
105.Huang W, Randhawa R, and Jain P, et al (2022) A novel artificial intelligence–powered method for prediction of early recurrence of prostate cancer after prostatectomy and cancer drivers JCO Clin Cancer Inform 6 131 https://doi.org/10.1200/cci.21.00131
106.Liu J, Zhang H, and Woon DTS, et al (2024) Predicting biochemical recurrence of prostate cancer post-prostatectomy using artificial intelligence: a systematic review Cancers (Basel) 16(21) 3596 https://doi.org/10.3390/cancers16213596 PMID: 39518036 PMCID: 11545810
107.Eminaga O, Saad F, and Tian Z, et al (2023) Artificial intelligence helps to predict recurrence and mortality for prostate cancer using histology images bioRxiv https://doi.org/10.1101/2023.07.27.550781
108.Pu L, Singha M, and Ramanujam J, et al (2022) CancerOmicsNet: a multi-omics network-based approach to anti-cancer drug profiling Oncotarget 13(1) 28234 https://doi.org/10.18632/oncotarget.28234
109.Gagare R, Sharma A, and Garg P (2023) AndroPred: an artificial intelligence-based model for predicting androgen receptor inhibitors J Biomolec Struct Dyn https://doi.org/10.1080/07391102.2023.2239935
110.Bosch D, Kuppen MCP, and Tascilar M, et al (2023) Reliability and efficiency of the CAPRI-3 metastatic prostate cancer registry driven by artificial intelligence Cancers 15(15) 3808 https://doi.org/10.3390/cancers15153808 PMID: 37568624 PMCID: 10417512
111.ClinicalTrials.gov National Library of Medicine (U.S.) (2000) Prospective Validation of Pathology-based Artificial Intelligence Diagnostic Model for Lymph Node Metastasis in Prostate Cancer Identifier NCT06253065 [https://clinicaltrials.gov/study/NCT06253065] Accessed date: 17/7/2024.
112.ClinicalTrials.gov National Library of Medicine (U.S.) (2000) AI-based Measurements of Tumour Burden in PSMA PET-CT Identifier NCT06363435 [https://clinicaltrials.gov/study/NCT06363435] Accessed date: 17/7/2024.
113.ClinicalTrials.gov National Library of Medicine (U.S.) (2000) Imperial Prostate 6 - Cancer Histology Artificial Intelligence Reliability Study (IP6-CHAIROS) Identifier NCT05228197 [https://clinicaltrials.gov/study/NCT05228197] Accessed date: 17/7/2024.
114.ClinicalTrials.gov National Library of Medicine (U.S.) (2000) Artificial Intelligence and Radiologists at Prostate Cancer Detection in MRI: The PI-CAI Challenge Identifier NCT05489341 [https://clinicaltrials.gov/study/NCT05489341] Accessed date: 17/7/2024.
115.ClinicalTrials.gov National Library of Medicine (U.S.) (2000) Accelerated Body Diffusion-Weighted MRI Using Artificial Intelligence (CeleScan-R) Identifier NCT05380609 [https://clinicaltrials.gov/study/NCT05380609] Accessed date: 17/7/2024.
116.ClinicalTrials.gov National Library of Medicine (U.S.) (2000) AI-Assisted System for Accurate Diagnosis and Prognosis of Breast Phyllodes Tumors Identifier NCT06286267 [https://clinicaltrials.gov/study/NCT06286267] Accessed date: 17/7/2024.
117.Xue S, Gafita A, and Dong C, et al (2022) Application of machine learning to pretherapeutically estimate dosimetry in men with advanced prostate cancer treated with 177Lu-PSMA I&T therapy Eur J Nucl Med Mol Imag 49(12) 4064–4072 https://doi.org/10.1007/s00259-022-05883-w
118.Deng K, Li H, and Guan Y (2020) Treatment stratification of patients with metastatic castration-resistant prostate cancer by machine learning iScience 23(2) 100804 https://doi.org/10.1016/j.isci.2019.100804 PMID: 31978751 PMCID: 6976944
119.Lin E, Hahn AW, and Nussenzveig RH, et al (2021) Identification of somatic gene signatures in circulating cell-free DNA associated with disease progression in metastatic prostate cancer by a novel machine learning platform Oncologist 26(9) 13869 https://doi.org/10.1002/onco.13869
120.Johnson KW, Torres Soto J, and Glicksberg BS, et al (2018) Artificial intelligence in cardiology J Am Coll Cardiol 71(23) 521 https://doi.org/10.1016/j.jacc.2018.03.521
121.Sun TQ and Medaglia R (2019) Mapping the challenges of artificial intelligence in the public sector: evidence from public healthcare Govern Inf Quart 36(2) 368–383 https://doi.org/10.1016/j.giq.2018.09.008
122.Jiang F, Jiang Y, and Zhi H, et al (2017) Artificial intelligence in healthcare: past, present and future Stroke Vasc Neurol 2(4) 101 https://doi.org/10.1136/svn-2017-000101
123.Bera K, Schalper KA, and Rimm DL, et al (2019) Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology Nat Rev Clin Oncol 16(11) 703–715 https://doi.org/10.1038/s41571-019-0252-y PMID: 31399699 PMCID: 6880861
124.Powles J and Hodson H (2017) Google deep mind and healthcare in an age of algorithms Health Technol 7(4) 351–367 https://doi.org/10.1007/s12553-017-0179-1
125.Dilsizian SE and Siegel EL (2014) Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment Curr Cardiol Rep 16(1) 441 https://doi.org/10.1007/s11886-013-0441-8
126.Luxton DD (2014) Recommendations for the ethical use and design of artificial intelligent care providers Artif Intell Med 62(1) 1–10 https://doi.org/10.1016/j.artmed.2014.06.004 PMID: 25059820
127.Hee Lee D and Yoon SN (2021) Application of artificial intelligence-based technologies in the healthcare industry: opportunities and challenges Int J Environ Res Public Health 18(1) 10271 https://doi.org/10.3390/ijerph18010271
128.Secinaro S, Calandra D, and Secinaro A, et al (2021) The role of artificial intelligence in healthcare: a structured literature review BMC Med Inform Decis Mak 21(1) 125 https://doi.org/10.1186/s12911-021-01488-9 PMID: 33836752 PMCID: 8035061
129.Caruana R, Lou Y, and Gehrke J, et al (2015) Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission Proc ACM SIGKDD Int Conf Know Discov Data Min 2015 1721–1730 https://doi.org/10.1145/2783258.2788613
130.Khan B, Fatima H, and Qureshi A, et al (2023) Drawbacks of artificial intelligence and their potential solutions in the healthcare sector Biomed Mater Dev 1(2) 63 https://doi.org/10.1007/s44174-023-00063-2
131.Laranjo L, Dunn AG, Tong HL, et al (2018) Conversational agents in healthcare: a systematic review J Am Med Inform Assoc 25(9) ocy072 https://doi.org/10.1093/jamia/ocy072
132.Topol EJ (2019) High-performance medicine: the convergence of human and artificial intelligence Nat Med 25(1) 44–56 https://doi.org/10.1038/s41591-018-0300-7 PMID: 30617339
133.Coiera E, Ammenwerth E, and Georgiou A, et al (2018) Does health informatics have a replication crisis? J Am Med Inform Assoc 25(8) ocy028 https://doi.org/10.1093/jamia/ocy028
134.Liu JLY and Wyatt JC (2011) The case for randomized controlled trials to assess the impact of clinical information systems J Am Med Inform Assoc 18(2) 10306 https://doi.org/10.1136/jamia.2010.010306
135.Coiera E and Tong HL (2021) Replication studies in the clinical decision support literature-frequency, fidelity, and impact J Am Med Inform Assoc 28(9) ocab049 https://doi.org/10.1093/jamia/ocab049
136.Spencer M (2015) Brittleness and bureaucracy: software as a material for science Perspect Sci 23(4) 184 https://doi.org/10.1162/POSC_a_00184
137.Kahn CE (2017) From images to actions: opportunities for artificial intelligence in radiology Radiology 285(3) 1734 https://doi.org/10.1148/radiol.2017171734
138.Wang Fand Preininger A (2019) AI in health: state of the art, challenges, and future directions Yearbook Med Inform 28(1) 908 https://doi.org/10.1055/s-0039-1677908
139.Nicholson Price II W (2017) Artificial intelligence in health care: applications and legal implications SciTech Lawyer 14 1
140.Watson DS, Krutzinna J, and Bruce IN, et al (2019) Clinical applications of machine learning algorithms: beyond the black box BMJ 364 l886 https://doi.org/10.1136/bmj.l886 PMID: 30862612
141.Williamson SM and Prybutok V (2024) Balancing privacy and progress: a review of privacy challenges, systemic oversight, and patient perceptions in AI-driven healthcare Appl Sci 14(2) 675 https://doi.org/10.3390/app14020675
142.Meskó B, Hetényi G, and Gyorffy Z (2018) Will artificial intelligence solve the human resource crisis in healthcare? BMC Health Serv Res 18(1) 545 https://doi.org/10.1186/s12913-018-3359-4 PMID: 30001717 PMCID: 6044098