Looking beyond the smokescreen: can the oral microbiome be a tool or target in the management of tobacco-associated oral cancer?
Snehal Kadam1, Madhusoodhanan Vandana1, Sudhanshu Patwardhan2 and Karishma S Kaushik1
1Human-Relevant Infection Biology Group, Institute of Bioinformatics and Biotechnology, Savitribai Phule Pune University, Pune 411007, India
2Centre for Health Research and Education, University of Southampton Science Park, Chilworth, Hampshire SO16 7NP, UK
Abstract
A wide range of microbes inhabit the oral cavity, and bacterial and fungal communities most often exist as structured communities or biofilms. The use of tobacco alters the structure of the oral microbiome, including that of potentially malignant lesions, and the altered oral microbiome influences key microenvironmental changes such as chronic inflammation, secretion of carcinogenic toxins, cellular and tissue remodelling and suppression of apoptosis. Given this, it is clear that the bacterial and fungal biofilms in potentially malignant states are likely not passive entities, but could play a critical role in shaping potential malignant and carcinogenic conditions. This holds potential towards leveraging the oral microbiome for the management of tobacco-associated potentially malignant lesions and oral cancer. Here, we explore this line of investigation by reviewing the effects of tobacco in shaping the oral microbiome, and analyse the available evidence in the light of the microbiome of oral potentially malignant and cancerous lesions, and the role of dysbiosis in carcinogenesis. Finally, we discuss possible interventions and approaches using which the oral microbiome could be leveraged towards precision-based oral cancer therapeutics.
Keywords: oral microbiome, biofilms, tobacco, potentially malignant lesions, oral cancer
Correspondence to: Karishma S Kaushik
Email: karishmaskaushik@gmail.com
Published: 01/02/2021
Received: 19/09/2020
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Tobacco use and oral cancer
Tobacco-associated disease is a global public health threat [1]. There are ~1.3 billion users of tobacco worldwide [2], of which an overwhelming majority (~80%) live in low- and middle-income countries. While a large proportion of tobacco users worldwide are cigarette smokers (~1 billion), the use of smokeless tobacco forms (nearly 30% in South and South-East Asia [3]) is also a grave concern. In India alone, there are ~300 million consumers of smokeless tobacco forms, and it is the most prevalent form of tobacco use among women in the country (India Report from CHRE [4]). Tobacco use is associated with a range of adverse health consequences [5, 6], notably the development of oral cancer [7].
Globally, tobacco use is responsible for 25% of all cancer-related deaths (2.4 million deaths from tobacco-use associated cancers) [8]. Cigarette smoke contains approximately 7,000 chemicals, of which at least 70 are known carcinogens [9–11]. Smoking-associated oral malignant changes are typically seen in the labial or buccal mucosa, tongue, gingiva, palate, alveolar mucosa, lips and salivary glands [12–15]. On the other hand, smokeless tobacco forms are chewed, placed in close contact with the oral mucosa or applied over the teeth and gums. Smokeless tobacco is consumed as raw leaves (local names such as khaini, misri, gutka, zarda, toombak) or dissolvable forms (lozenges, sticks, strips) [3, 16], often containing additives such as areca nut, betel quid, catechu, slaked lime, ash, sodium bicarbonate, as well as flavouring agents such as menthol and plant oils. The process of tobacco curing, fermentation and ageing, results in the production of tobacco-specific nitrosamines (TSNAs), which are the major group of carcinogens in smokeless tobacco [17–19].
Current management of tobacco-associated premalignant changes and oral cancer
Tobacco-associated oral cancer typically starts as premalignant lesions, which through a series of genetic and molecular changes, influenced by oral microenvironmental factors [20, 21], undergoes malignant transformation to oral cancer. The most common form of oral cancer squamous cell carcinoma [22], which most often develops from potentially malignant lesions such as oral submucous fibrosis, leukoplakia, erythroplakia, among others [23, 24].
Currently, the mainstay of management for oral premalignant lesions is observation with frequent clinical examinations, along with education towards cessation. This is not only a passive and suboptimal approach, but is also limited by access to healthcare and success of cessation efforts. On the other hand, active interventional approaches include surgical resection and ablation. These are limited by ‘field effects’ where potentially malignant changes, though not phenotypically visible, extend to larger areas beyond the lesion, as well as severe disfigurement and impairment of function. Finally, limited medical approaches such as retinoic acid, Vitamin E, and natural compounds are either limited by toxicity (retinoic acid) or lack of thorough clinical evaluations [20]. As evident, there is an impasse in the management of tobacco-associated potentially malignant lesions, and current management strategies fail to account for the complexity of the disease state.
Tobacco, the oral microbiome and oral cancer
Tobacco-associated potentially malignant lesions exist and develop in a dynamic oral microenvironment, that includes diverse, multi-species microbial communities [25–28], most often observed as biofilms. It is widely recognised that distinct microbial signatures influence key processes such as chronic inflammation and carcinogenesis [29, 30]. This offers the exciting possibility that the oral microbiome (or biofilms) can be a tool or target in the management of tobacco-associated potentially malignant lesions. Here, we explore this line of investigation by reviewing the increasingly recognized role of tobacco in shaping the oral microbiome, focusing on the bacterial and fungal communities (Table 1). We then look at the effects of bacterial and fungal dysbiosis on key processes in oral potentially malignant and cancerous lesions. Finally, we discuss possible interventions and approaches with which the oral microbiome can be leveraged towards precision-based oral cancer therapeutics.
How tobacco affects the oral microbiome
Tobacco is known to influence the inflammatory response of the oral cavity [31–42]. Overall, this contributes to a pro-inflammatory microenvironment eventually leading to chronic inflammation, which is known to be associated with oral carcinogenesis [43–47]. However, it is evident that tobacco exposure (in various forms) not only results in carcinogenic changes in the oral mucosa [48–57], but also shapes the oral microbiome. Several studies have characterized the oral microbiome in tobacco users [36, 58–64], using samples such as tumour or lesion biopsies, oral biofilm swabs, oral cavity rinses or saliva, from a diverse range of tobacco users.
Table 1. Tobacco-associated oral microbiome changes associated with the development and progression of premalignant lesions and oral cancer.
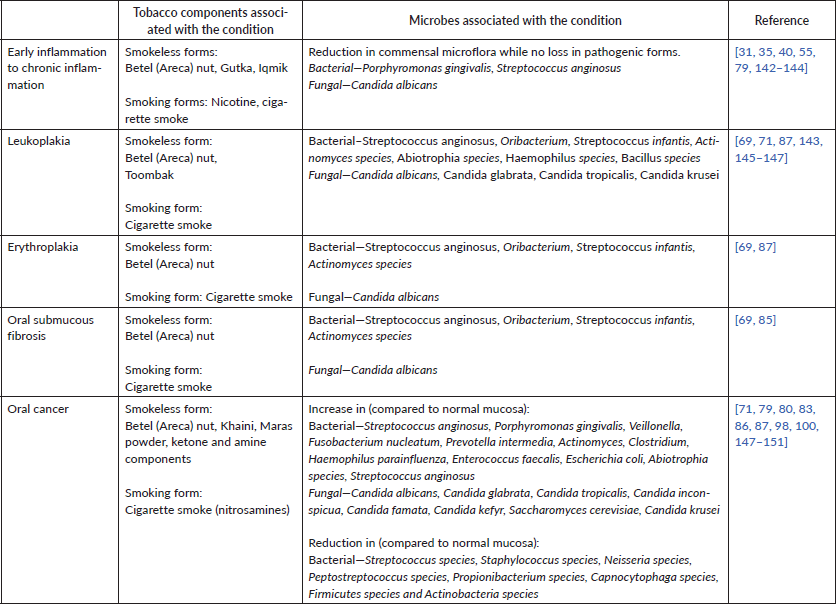
Alteration in bacterial species profiles
In one study, tobacco smokers and non-smokers were sampled for oral marginal and subgingival plaques [36]. During this study, all subjects received oral prophylaxis to remove any initial plaque and were asked to cover six teeth while brushing to avoid loss of the plaque. After 24 hours, the plaque samples were collected and the procedure was repeated to obtain samples at different time points up to 7 days. This model allowed the study of undisturbed plaque from subjects for the given time periods. About 71% of the oral plaque community in non-smokers remained stable over the 7-day period, whereas this number was only 46% in smokers, indicating greater fluctuations in smoker plaques. Over a period of 7 days, smokers acquired species like Lactobacillus, Fusobacterium, Centipeda periodontii, Pseudomonas, Treponema, Leptotrichia, Synergistes, Propionibacterium and Cardiobacterium, which were absent in non-smokers [36]. In a similar study, tobacco smoking contributed to a pathogen-rich environment as seen by the presence of species such as F. nucleatum, F. naviforme, A. johnsonii, A. baumannii, A. haemolyticus, S. mutans, and low abundance of Streptococcus sanguinis, S. oralis, Actinomyces viscosus, A. israelii, A. dentalis, Neisseria subflava [63, 64].
In another study of smokers and non-smokers, with further subgroups of treated and non-treated for periodontitis, Fusobacterium was found to be higher in both untreated and treated smoker groups [65]. Additionally, the same study found Bacteroides forsythus, Peptostreptococcus micros and Campylobacter rectus, along with Fusobacterium nucleatum, to be the most prominent microbial signature for smokers. Though studies have noted an increase in Fusobacteria in smoker populations, some studies report an opposite trend [59, 66, 67]. Interestingly, under in vitro conditions, smokeless tobacco aqueous extracts and TSNAs demonstrated either a reduction of growth for Fusobacterium nucleatum or no change in profile. All smokeless extracts promoted the growth of Peptostreptococcus micros, Streptococcus anginosus, S. constellatus, S. sanguinis and Veillonella parvula [68]. For certain species, this also correlates to what is seen in vivo, where Streptococcus anginosus was significantly higher in chewers having oral lesions [69], and has been identified in various head and neck cancers [70–72]. Notably, Streptococcus infantis was more abundant in current chewers compared to past users or those who never used the tobacco product [69], indicating that some differences in microbiome due to betel quid chewing could be reversible.
On the other hand, clinical saliva samples of tobacco smokers revealed reduced diversity of Gram-positive bacteria compared to non-smokers, with only nine different species cultured from smokers compared to eighteen different species from non-smokers [61]. Certain Proteobacteria were also reduced in smokers [59, 60, 62].
Alteration in bacterial gene expression profiles
In addition to altered species profiles in tobacco users, tobacco smoke exposure elicits different gene expression profiles in multi-species pathogenic and commensal biofilms [35]. Commensal biofilms were grown by seeding Streptococcus oralis, Streptococcus sanguis, Streptococcus mitis, Veillonella parvula, Neisseria mucosa, and Actinomyces naeslundii in a mixture of Brain -Heart Infusion broth (BHI) and artificial saliva (1:1). Pathogenic biofilms were grown by seeding commensals along with F. nucleatum, and incubating for 24 hours, followed by further seeding of Porphyromonas gingivalis, Filifactor alocis, Dialister pneumosintes, Selenonomas sputigena, Selenominas noxia, Catonella morbi, Parvimonas micra and Tannerella forsythia. Smoke exposure upregulated fermentative pathways in both pathogenic and commensal biofilms. Fermentative pathways among several bacteria can lead to the formation of metabolites like butyrate [73], and the role of butyrate has been studied in various cancers [74, 75]. Commensal biofilms in tobacco smoke environments faced a loss of viability and elicited an early proinflammatory immune response (seen as an increase in various interleukins and macrophage inflammatory proteins) from immortalised human oral keratinocytes (OKF6/TERT-2) [35]. Pathogenic biofilms, however, did not show any significant loss of viability during the same time period, but did elicit a late immune response. This could indicate that tobacco exposure leads to elimination of commensal biofilms, while selecting for pathogenic biofilms. This transition from a commensal dominant to a pathogen dominant biofilm with tobacco usage could serve as a marker for oral health decline and increased risks of lesion progression.
Effect on bacterial interspecies interactions
When Porphyromonas gingivalis grown in cigarette smoke extract-conditioned media (compared to non-conditioned media) was added on top of Streptococcus gordonii on saliva-coated coverslips in flow cells, a three-fold increase in the dual-species biofilm height and two-fold increase in total microcolony numbers were detected [76]. Peripheral blood mononuclear cells challenged with cigarette smoke extract-exposed P. gingivalis biofilms (on pellicle coated discs) exhibited lower Interleukin (IL)-6 and Tumor Necrosis Factor (TNF)-α, compared to control biofilms. Cigarette smoke extract exposure was thus seen to promote dual-species biofilms of P. gingivalis and S. gordonii, and lowers the proinflammatory response elicited by P. gingivalis. Cigarette smoke extract-exposure increased P. gingivalis binding to immobilised Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a FimA ligand on S. gordonii. a FimA ligand on S. gordonii. Additionally, cigarette smoke extract-exposure was shown to increase FimA protein levels, an important virulence factor that helps in adhesion and attachment, and also increases biofilm formation by P. gingivalis [77]. A recent study has shown that P. gingivalis invaded OKF6/TERT cells less in the presence of commensals such as S. gordonii [78]. An increase in dual-biofilm formation upon cigarette smoke extract-exposure suggests that tobacco could possibly alter the interactions between the two bacterial species allowing pathogens to use existing commensals to establish themselves in the oral cavity. Various studies have isolated P. gingivalis from oral tumours [79, 80], and infection with P. gingivalis resulted in increased tongue lesion size in a 4-nitroquinoline 1-oxide (4NQO) based cancer mouse model [81]. Moreover, infection with P. gingivalis along with 4-quinoline oxide (QO), a carcinogen, led 65% of the mice to develop squamous cell carcinoma compared to 45% in mice treated with only 4NQO, indicating that P. gingivalis enhanced carcinogenesis. P. gingivalis also showed increased staining in gingival carcinomas as compared to S. gordonii [80], supporting the non-invasiveness of S. gordonii with respect to gingival epithelial cells in vitro [82].
Effect on fungal species profiles
Among fungal species, Candida albicans and non-albicans Candida species such as C. tropicalis, C. krusei, C. glabrata have been isolated from various oral lesions [83–87]. C. albicans was significantly higher in oral swabs of waterpipe smokers compared to non-smokers [66]. Saliva samples from tobacco users had higher levels of Candida albicans and C. tropicalis and C. krusei was unchanged compared to non-tobacco users [88]. This trend was also observed with smokeless tobacco users (gutka and betel-quid chewers), who had a higher Candida carriage, compared to non-chewers, with C. albicans being the most commonly present species [89].
Different concentrations of cigarette smoke condensate have been shown to result in increased adhesion on glass slides as well as increased metabolic activity in cultures of C. albicans [90]. C. albicans also formed more biofilms on collagen scaffolds, as seen by scanning electron microscopy as well as crystal violet staining. This increased adhesion and biofilm formation of Candida biofilms in presence of cigarette smoke condensate has also been noted for orthodontic materials like acrylic resin and bands [91]. Cigarette smoke condensate (CSC), in the presence of 10% serum at 37°C (hyphae-forming conditions), was observed to increase C. albicans in vitro hyphal formation and chitin production [92]. C. albicans pretreated with CSC were not only able to adhere more to human gingival fibroblasts, but also proliferated more within 48 hours. A higher number of CSC-pretreated Candida cells transitioned to hyphal form upon contact with the fibroblasts under hyphal specific conditions compared to those not treated with CSC. Attachment to host cells and increased hyphal formation are important for the invasiveness of Candida [93], with Candida species isolated from oral mucosal lesions associated with tobacco use were found to be in their hyphal forms [85].
So far, the evidence strongly establishes that tobacco use alters the distribution, diversity and abundance of oral bacterial and fungal species, relative to non-users. The fact that the studies vary in their findings, underscores that various factors such as tobacco type, usage pattern, duration and frequency, underlying predisposing factors, health conditions and diet, play an additional role [94]. Notwithstanding this, altered oral microbial communities resulting from tobacco use are very likely to shape the microbiome of tobacco-associated potentially malignant lesions.
Microbes associated with oral potentially malignant lesions and cancer
Microbes, often as biofilms, have been found in close association with potentially malignant lesions and Oral Squamous Cell Carcinoma (OSCC) tumours in the oral cavity [86, 95–101]. Notably, the abundance, distribution, diversity have been observed to vary not only between tobacco-users and non-users, but also across tumour and non-tumour sites in OSCC patients [98, 102, 103]. Bacterial communities on the surface of oral carcinomas in tobacco-users have to be enriched for Fusobacterium, Actinomyces, Clostridium, Enterobacteriaceae and Streptococcus species, Klebsiella pneumoniae, Enterococcus faecalis, Veillonella, Prevotella, Porphyromonas and Clostridium [98, 102, 103]. In one study, three groups, comprising of OSCC tumour patients, high-risk individuals (smokers and alcohol users without signs of OSCC) and healthy individuals, were analysed for microbial species and abundance (either at tumour sites of OSCC subjects or normal mucosa in high-risk and healthy subjects) [100]. Patients with OSCC tumours showed a higher presence of anaerobes, relative to the other two groups, with Fusobacterium and Prevotella seen to predominate. Another study identified a large cohort of bacteria within OSCC tissues, as visualised by fluorescent in situ hybridisation [96]. Bacteria were found to be present throughout the OSCC tissue. Bacterial identification via 16S rRNA sequencing revealed that Clavibacter michiganensis subsp. tessellarius, Fusobacterium naviforme and Ralstonia insidiosa were at least 30% more in OSCC samples. Streptococcus anginosus was also identified in a subset of patient in this study. Notably, Streptococcus anginosus has been found in close association with many head and neck carcinomas [70, 71]. Notably, S. anginosus isolated from OSCC samples was genotypically identical to S. anginosus isolated from dental plaques of the same patients, and was not detected in saliva samples, indicating that plaques could be locations where the bacteria accumulate in the oral cavity [71].
Candida species have been isolated from patients having malignant oral squamous cell carcinoma, atypical lichen planus and chronic candidiasis, and Candida species from dorsum tongue of asymptomatic carriers with healthy mucosa were used as controls [83]. Candida albicans was the most commonly occurring species in all groups. Candida albicans strains of different biotypes isolated from leukoplakia and erythroleukoplakia have demonstrated high nitrosation potential, quantified as the ability to form N-nitroso benzylmethylamine from N-benzylmethylamine and nitrite [104]. Higher nitrosation potential isolates correlated with advanced stages of lesions. Other Candida species, such as C. tropicalis and C. parapsilosis, isolated from these lesions ranked lower in their nitrosation potential. The ability to produce carcinogens like nitrosamines has been suggested to contribute to malignant transformation in the oral cavity [105–107]. Further, in vitro analysis revealed that isolates from the carcinoma and candidiasis groups were able to form significantly more biofilms as quantified by metabolic activity (XTT); notably, Candida albicans formed less biofilms compared to other strains. On the other hand, no difference was found in lipolytic activity, proteolytic activity and hydrophobicity. Though the virulence factors had differential levels in different clinical conditions, there was no association of virulence factors with individual strain types. This indicates that Candida pathogenesis is likely to be influenced by an interplay of multiple factors like virulence, abundance of species, as well as other underlying factors like the immune state of the patient and co-infecting species.
In a recent study, the diversity and distribution of microbial signatures was largely seen to reduce with cancer progression. When samples of normal, epithelial precursor lesions (dysplasia, hyperplasia or hyperkeratosis) and OSCC individuals were analysed for microbial composition, a multidimensional scaling plot revealed that the distribution was more condensed for OSCC patients than for the other two groups [103]. Harvesting of tissue samples from oral tumours and 16S rRNA sequencing based identification of microbial isolates revealed that the microbiome of superficial tissues of the tumour housed additional species which were absent in the deep tumour tissues [108]. This indicates a specialisation within the tumour itself, where not all species in the oral cavity would become associated with and survive in the tumour microenvironment.
Taken together, certain key microbial players are observed to be consistently detected in association with oral potentially malignant lesions and OSCC, including Fusobacterium, Candida, Porphyromonas, Streptococcus, Veillonella and Prevotella. While evidence is limited in this regard, it is likely that the microbial profile changes with lesion progression, pointing to the imminent role of the microbiome in shaping inflammatory and carcinogenic processes.
Oral dysbiosis and cancer: how could the altered oral microbiome play a role in carcinogenesis?
Microbial dysbiosis is well-known to be associated with carcinogenic processes [109–112], either via inflammatory changes, or the production of toxins or metabolites. It is likely that these processes play out in potentially malignant and OSCC lesions, and could initiate and promote a series of inflammatory and potentially carcinogenic changes in the oral cavity. In general, multiple oral species, via secreted endotoxins and metabolic byproducts, induce production of pro-inflammatory cytokines (such as TNFɑ, IL-1, IL-6, IL-8), other immune signaling factors (such as Matrix Metallo-Proteinases (MMPs), Granulocye-Macrophage Colony-Stimulating Factor (GM-CSF)), degradation of tissues, inhibition of antibacterial activities of immune cells and invasion of host tissues [26, 113–116]. This results in a chronic inflammatory microenvironment, known to affect key carcinogenic processes such as cell growth, proliferation and migration, apoptosis and differentiation to tumour-like phenotypes [105, 113, 117–119]. In the context of oral carcinogenesis, notable pathogens include Fusobacterium nucleatum, Porphyromonas gingivalis and Candida species.
Porphyromonas gingivalis and Fusobacterium nucleatum produce sulphur compounds that induce cell proliferation, migration, invasion and tumour angiogenesis. In vitro infection of an oral cancerous cell line (OQ01) with Fusobacterium nucleatum showed increased IL-8 production and invasiveness of the cell line [120]. In another in vitro study, human OSCC cells showed increased proliferation and invasiveness when co-incubated with P. gingivalis and F. nucleatum. Prolonged exposure resulted in a change in OSCC morphology from polygonal to elongated, a decrease in the levels of the epithelial marker Cytokeratin-13 and an increase of mesenchymal markers (N-cadherin and α-Smooth Muscle Acton (SMA)), indicating an Epithelial-to-Mesemchymal Transition (EMT) phenotype [121]. P. gingivalis has also been shown to inhibit apoptosis, and enhance cell proliferation and cellular invasion with F. nucleatum [122–129], and their role in promoting OSCC has been characterised extensively [26, 99, 130]. In an in vivo 4NQO-induced carcinoma model, mice administered with 4NQO and co-infected with both P. gingivalis and F. nucleatum developed tongue tumours 2.5 times in size compared to mice only administered 4NQO and had an increased expression of the oncogene cyclin D1 [131].
Candida species have been widely reported in association with oral potentially malignant lesions and in OSCC samples. Through hyphal formation, that enables active penetration or endocytosis, Candida species have been shown to invade epithelial cells [132–136]. Further contact-sensing and hyphal extension, combined with secretion of toxins and virulence factors induces damage to the oral epithelia [137–139]. Candida species also produce nitrosamines and their nitrosation potential can lead to DNA damage, and eventually initiate carcinogenic changes [107, 140]. Candida species have been isolated from lesions of oral leukoplakia, where a higher nitrosation potential of the fungal strain was associated with an advanced potentially malignant stage [104]. Therefore, it is likely that increased Candida abundance, adhesion and invasion with tobacco usage, and the potential of Candida-induced nitrosative changes plays a role in the development of potentially malignant lesions and progression to cancer [139, 141].
Leveraging the oral microbiome as a tool or target in the management of tobacco-associated oral potentially malignant lesions and cancer
Taken together, the use of tobacco alters the oral microbiome, and the oral microbiome influences key pathways involved in inflammation and carcinogenesis (Figure 1). Given this, it is clear that the oral microbiome in potentially malignant states plays a critical role in shaping the oral carcinogenic microenvironment. Therefore, leveraging the oral microbiome towards the management of tobacco-associated potentially malignant lesions and oral cancer could hold potential. For this, there are several strategies and approaches that could be adopted.
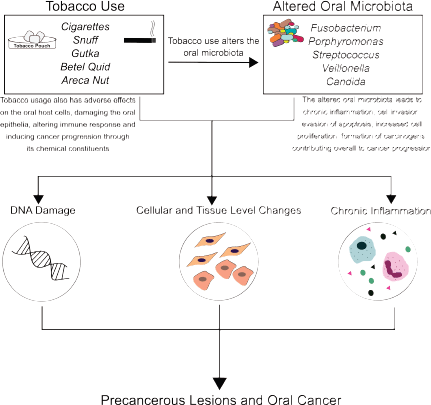
Figure 1. Role of tobacco and microbes in oral carcinogenesis: Tobacco use, its forms, products and additives (cigarette, snuff, gutka, betel quid, areca nut), and derivatives (like tobacco-specific nitrosamines) have direct carcinogenic effects on the oral mucosa. Tobacco use also alters the oral microbiome, resulting in distinct microbial signatures in normal mucosa, potentially malignant lesions and cancerous tissue. Together, this altered microbiome and tobacco use result in a proinflammatory microenvironment, contributing to potentially malignant lesions, and oral carcinogenesis.
Profiling the oral microbiome in tobacco-associated potentially malignant lesions: Using a combination of standard microbiology approaches and molecular tools, the biogeography of the oral microbiome in a range of tobacco-associated clinical conditions, across variations in diet, lifestyle, associated risk factors, can be profiled. This will serve as a baseline for screening and interventional tools in local populations, and is particularly important given the variations in the microbial profile across study conditions.
Probing the microbiome of tobacco-associated potentially malignant lesions as a cancer screening tool: Based on previous studies, the progression of potentially malignant lesions to oral cancer is associated with an alteration in the microbiome, and levels of microbial metabolites. Detection of these ‘signature’ markers in potentially malignant lesions could be leveraged as screening approaches. This would serve as a valuable addition to macroscopic (visual) screening for cancerous changes, and be less invasive as compared with routine biopsies. Most importantly, identifying microbial changes early in cancer progression would provide the possibility of targeting the oral microbiome, an active interventional approach to preventing progression to oral cancer.
Targeting the oral microbiome in the management of potentially malignant lesions and oral cancer: For this, a wide range of antimicrobial approaches, including conventional antibiotics and antifungal agents, can be employed. However, given the rise in antibiotic resistance, novel antimicrobial approaches such as peptides, natural extracts, polyphenols, nucleic-acid mimics and structural analogues can be evaluated for antimicrobial and antibiofilm effects. Additional approaches could include targeting the proinflammatory markers, including cytokines and growth factors, known to be dysregulated in potentially malignant lesions, and cancer progression states.
Conclusion
In conclusion, exploring interactions between tobacco use, the oral microbiome and carcinogenesis could not only lead to identification of potential biomarkers, but also lead to precision-treatment approaches that shape the oral microbiome towards a better therapeutic outcomes for oral potentially malignant lesions and cancer. This will provide comprehensive insights into the ‘microbiome paradigm’ for the management of tobacco-associated oral carcinogenesis, and open unexplored lines of investigation into precision-based oral cancer therapeutics.
Conflicts of interest
Dr Sudhanshu Patwardhan (SP) is a paid Director for Centre for Health Research and Education (CHRE) UK, an independent healthcare company, which works on projects on smoking cessation and cancer prevention globally. CHRE has received grants from Foundation for a Smoke-free World, Inc for some of its smoking cessation projects. For this article, SP or CHRE did not receive any external funding.
Funding statement
The academic appointments of Karishma S Kaushik (KSK) and Snehal Kadam are funded via the Ramalingaswami Re-entry Fellowship (to KSK), Department of Biotechnology, Government of India. This review was commissioned and funded by CHRE.
Acknowledgments
The academic appointments of Karishma S Kaushik and Snehal Kadam are funded via the Ramalingaswami Re-entry Fellowship (to KSK).
References
1. Wipfli H and Samet JM (2016) One hundred years in the making: the global tobacco epidemic Annu Rev Public Heal 37 149–166 https://doi.org/10.1146/annurev-publhealth-032315-021850
2. WHO launches new report on global tobacco use trends (n.d.) [https://www.who.int/news-room/detail/19-12-2019-who-launches-new-report-on-global-tobacco-use-trends] Date accessed: 12/08/20
3. Siddiqi K, Shah S, and Abbas SM, et al (2015) Global burden of disease due to smokeless tobacco consumption in adults: analysis of data from 113 countries BMC Med 13 194 https://doi.org/10.1186/s12916-015-0424-2 PMID: 26278072 PMCID: 4538761
4. The Foundation for a Smoke-Free World. India Country Report 2020
5. Gupta PC (2001) The public health impact of tobacco Curr Sci 81 475–481 https://doi.org/10.2307/24105879
6. Bonnie RJ, Stratton K, and Kwan LY (2015) Products C on the PHI of R the MA for PT, Practice B on PH and PH, Medicine I of. The Effects of Tobacco Use on Health
7. Khani Y, Pourgholam-Amiji N, and Afshar M, et al (2018) Tobacco smoking and cancer types: a review Biomed Res Ther 5 2142–2159 https://doi.org/10.15419/bmrat.v5i4.428
8. WHO (2020) Who report on cancer: setting priorities, investing wisely and providing care for all
9. Surgeon General (2010) How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease (Atlanta, GA: Centers for Disease Control and Prevention)
10. Campain JA (2004) Nicotine: potentially a multifunctional carcinogen? Toxicol Sci 79(1) 1–3 https://doi.org/10.1093/toxsci/kfh106 PMID: 15146885
11. Smith CJ, Livingston SD, and Doolittle DJ (1997) An international literature survey of “IARC group I carcinogens” reported in mainstream cigarette smoke Food Chem Toxicol 35 1107–1130 https://doi.org/10.1016/S0278-6915(97)00063-X
12. Perry BJ, Zammit AP, and Lewowski AW, et al (2020) Sites of origin of oral cavity cancer in nonsmokers vs smokers: possible evidence of dental trauma carcinogenesis and its importance compared with human papillomavirus JAMA Otolaryngol - Head Neck Surg 141 5–11 https://doi.org/10.1001/jamaoto.2014.2620
13. Dhanuthai K, Rojanawatsirivej S, and Thosaporn W, et al (2018) Oral cancer: a multicenter study Med Oral Patol Oral Cir Bucal 23 e23–e29 https://doi.org/10.4317/medoral.21999 PMCID: 5822535
14. Morse DE, Psoter WJ, and Cleveland D, et al (2007) Smoking and drinking in relation to oral cancer and oral epithelial dysplasia Cancer Causes Control 18 919–929 https://doi.org/10.1007/s10552-007-9026-4 PMID: 17647085 PMCID: 2139900
15. Location of Oral Cancers Differs in Smokers, Nonsmokers - For The Media - JAMA Network n.d. [https://media.jamanetwork.com/news-item/location-of-oral-cancers-differs-in-smokers-nonsmokers/] Date accessed: 2/08/20
16. Johnson N (2001) Tobacco use and oral cancer: a global perspective J Dent Educ 65(4) 328–339 https://doi.org/10.1002/j.0022-0337.2001.65.4.tb03403.x PMID: 11336118
17. Warnakulasuriya S and Straif K (2018) Carcinogenicity of smokeless tobacco: evidence from studies in humans & experimental animals Indian J Med Res 148 681–686 https://doi.org/10.4103/ijmr.IJMR_149_18
18. Hoffmann D, Rivenson A, and Chung FL, et al (1991) Nicotine-derived n-nitrosamines (TSNA) and their relevance in tobacco carcinogenesis Crit Rev Toxicol 21 305–311 https://doi.org/10.3109/10408449109017917 PMID: 2069715
19. Preston-Martin S (1991) Evaluation of the evidence that tobacco-specific nitrosamines (TSNA) cause cancer in humans Crit Rev Toxicol 21 295–298 https://doi.org/10.3109/10408449109017915 PMID: 2069713
20. Gillenwater A, Papadimitrakopoulou V, and Richards-Kortum R (2006) Oral premalignancy: New methods of detection and treatment Curr Oncol Rep 8 146–154 https://doi.org/10.1007/s11912-006-0050-4 PMID: 16507225 PMCID: 2773158
21. Peltanova B, Raudenska M, and Masarik M (2019) Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review Mol Cancer 18 1–24 https://doi.org/10.1186/s12943-019-0983-5
22. Rivera C (2015) Essentials of oral cancer Int J Clin Exp Pathol 8 11884–11894 PMID: 26617944 PMCID: 4637760
23. Ganesh D, Sreenivasan P, and Ohman J, et al (2018) Potentially malignant oral disorders and cancer transformation Anticancer Res 38 3223–3229 https://doi.org/10.21873/anticanres.12587 PMID: 29848669
24. Warnakulasuriya S, Johnson NW, and Van Der Waal I (2007) Nomenclature and classification of potentially malignant disorders of the oral mucosa J Oral Pathol Med 36 575–580 https://doi.org/10.1111/j.1600-0714.2007.00582.x PMID: 17944749
25. Lamont RJ, Koo H, and Hajishengallis G (2018) The oral microbiota: dynamic communities and host interactions Nat Rev Microbiol 16 745–759 https://doi.org/10.1038/s41579-018-0089-x PMID: 30301974 PMCID: 6278837
26. Chattopadhyay I, Verma M, and Panda M (2019) Role of oral microbiome signatures in diagnosis and prognosis of oral cancer Technol Cancer Res Treat 18 https://doi.org/10.1177/1533033819867354 PMID: 31370775 PMCID: 6676258
27. Avila M, Ojcius DM, and Yilmaz Ö (2009) The oral microbiota: living with a permanent guest DNA Cell Biol 28 405–411 https://doi.org/10.1089/dna.2009.0874 PMID: 19485767 PMCID: 2768665
28. Chen T, Yu WH, and Izard J, et al (2010) The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information Database (Oxford) 2010 baqo13 https://doi.org/10.1093/database/baq013
29. Srinivasprasad V, Dineshshankar J, and Sathiyajeeva J, et al (2015) Liaison between micro-organisms and oral cancer J Pharm Bioallied Sci 7 S354–S360 https://doi.org/10.4103/0975-7406.163451 PMID: 26538877 PMCID: 4606619
30. Whitmore SE and Lamont RJ (2014) Oral bacteria and cancer PLoS Pathog 10 https://doi.org/10.1371/journal.ppat.1003933 PMID: 24676390 PMCID: 3968118
31. Johnson GK, Guthmiller JM, and Joly S, et al (2010) Interleukin-1 and interleukin-8 in nicotine- and lipopolysaccharide-exposed gingival keratinocyte cultures J Periodontal Res 45 583–588 https://doi.org/10.1111/j.1600-0765.2009.01262.x PMID: 20337880
32. Johnson GK, Poore TK, and Squier CA, et al (1994) Prostaglandin E2 and interleukin‐1 levels in smokeless tobacco‐induced oral mucosal lesions J Periodontal Res 29 430–438 https://doi.org/10.1111/j.1600-0765.1994.tb01245.x PMID: 7877079
33. Upadhyay J, Upadhyay RB, and Agrawal P, et al (2013) Langerhans cells and their role in oral mucosal diseases N Am J Med Sci 5 505–514 https://doi.org/10.4103/1947-2714.118923 PMID: 24251267 PMCID: 3818822
34. Daniels TE, Chou L, and Greenspan JS, et al (1992) Reduction of Langerhans cells in smokeless tobacco-associated oral mucosal lesions J Oral Pathol Med 21 100–104 https://doi.org/10.1111/j.1600-0714.1992.tb00990.x PMID: 1374796
35. Shah SA, Ganesan SM, and Varadharaj S, et al (2017) The making of a miscreant: Tobacco smoke and the creation of pathogen-rich biofilms Npj Biofilms Microbiomes 3(1) https://doi.org/10.1038/s41522-017-0033-2 PMID: 29081982 PMCID: 5655325
36. Kumar PS, Matthews CR, and Joshi V, et al (2011) Tobacco smoking affects bacterial acquisition and colonization in oral biofilms Infect Immun 79 4730–4738 https://doi.org/10.1128/IAI.05371-11 PMID: 21859855 PMCID: 3257914
37. Pabst MJ, Pabst KM, and Collier JA, et al (1995) Inhibition of neutrophil and monocyte defensive functions by nicotine J Periodontol 66 1047–1055 https://doi.org/10.1902/jop.1995.66.12.1047 PMID: 8683417
38. Rodríguez-Rabassa M, López P, and Rodríguez-Santiago RE, et al (2018) Cigarette smoking modulation of saliva microbial composition and cytokine levels Int J Environ Res Public Health 15(11) 2479 https://doi.org/10.3390/ijerph15112479 PMCID: 6266915
39. Lee J, Taneja V, and Vassallo R (2012) Cigarette smoking and inflammation: cellular and molecular mechanisms J Dent Res 91 142–149 https://doi.org/10.1177/0022034511421200 PMCID: 3261116
40. Javed F, Al-Kheraif AA, and Al Amri MD, et al (2015) Periodontal parameters and whole salivary cytokine profiles among habitual gutka chewers and non-chewers J Periodontol 86 689–695 https://doi.org/10.1902/jop.2015.140556 PMID: 25612632
41. Johnson GK, Payne JB, and Fili JM, et al (2007) Development of smokeless tobacco-induced oral mucosal lesions J Oral Pathol Med 27 388–394 https://doi.org/10.1111/j.1600-0714.1998.tb01972.x
42. Sundar IK, Javed F, and Romanos GE, et al (2016) E-cigarettes and flavorings induce inflammatory and pro-senescence responses in oral epithelial cells and periodontal fibroblasts Oncotarget 7 77196–77204 https://doi.org/10.18632/oncotarget.12857 PMID: 27791204 PMCID: 5363580
43. Deepthi G, Nandan SRK, and Kulkarni PG (2019) Salivary tumour necrosis factor-α as a biomarker in oral leukoplakia and oral squamous cell carcinoma Asian Pacific J Cancer Prev 20 2087–2093 https://doi.org/10.31557/APJCP.2019.20.7.2087
44. Ameena M and Rathy R (2019) Evaluation of tumor necrosis factor: Alpha in the saliva of oral cancer, leukoplakia, and healthy controls – a comparative study J Int Oral Heal 11 92 https://doi.org/10.4103/jioh.jioh_202_18
45. Hoare A, Soto C, and Rojas-Celis V, et al (2019) Chronic inflammation as a link between periodontitis and carcinogenesis Mediators Inflamm2019 1029857 https://doi.org/10.1155/2019/1029857
46. Lee CH, Chang JSM, and Syu SH, et al (2015) IL-1β promotes malignant transformation and tumor aggressiveness in oral cancer J Cell Physiol 230 875–884 https://doi.org/10.1002/jcp.24816
47. Seyedroudbari SA and Khan MM (1998) In vitro effects of smokeless tobacco extract on tumor necrosis factor- α (TNF-α) and interleukin-1β (IL-1β) production, and on lymphocyte proliferation Toxicon 36 631–637 https://doi.org/10.1016/S0041-0101(97)00092-5 PMID: 9643476
48. Ma B, Stepanov I, and Hecht SS (2019) Recent studies on DNA adducts resulting from human exposure to tobacco smoke Toxics 7(1) 16 https://doi.org/10.3390/toxics7010016 PMCID: 6468371
49. Pryor WA (1997) Cigarette smoke radicals and the role of free radicals in chemical carcinogenicity Environ Health Perspect 105(Suppl 4) 875–882 https://doi.org/10.1289/ehp.97105s4875 PMID: 9255574 PMCID: 1470037
50. Valavanidis A, Vlachogianni T, and Fiotakis K (2009) Tobacco smoke: Involvement of reactive oxygen species and stable free radicals in mechanisms of oxidative damage, carcinogenesis and synergistic effects with other respirable particles Int J Environ Res Public Health 6 445–462 https://doi.org/10.3390/ijerph6020445 PMID: 19440393 PMCID: 2672368
51. Lipinski B (2011) Hydroxyl radical and its scavengers in health and disease Oxid Med Cell Longev 2011 809696 https://doi.org/10.1155/2011/809696 PMID: 21904647 PMCID: 3166784
52. Nair UJ, Floyd RA, and Nair J, et al (1987) Formation of reactive oxygen species and of 8-hydroxydeoxyguanosine in DNA in vitro with betel quid ingredients Chem Biol Interact 63 157–169 https://doi.org/10.1016/0009-2797(87)90095-0 PMID: 3664791
53. Burney S, Caulfield JL, and Niles JC, et al (1999) The chemistry of DNA damage from nitric oxide and peroxynitrite Mutat Res - Fundam Mol Mech Mutagen 424 37–49 https://doi.org/10.1016/S0027-5107(99)00006-8
54. Yalcin E and de la Monte S (2016) Tobacco nitrosamines as culprits in disease: mechanisms reviewed J Physiol Biochem 72 107–120 https://doi.org/10.1007/s13105-016-0465-9 PMID: 26767836 PMCID: 4868960
55. Pappas RS, Stanfill SB, and Watson CH, et al (2008) Analysis of toxic metals in commercial moist snuff and Alaskan Iqmik 32(4) 281–291 https://doi.org/10.1093/JAT/32.4.281 PMID: 18430295
56. Bhat MY, Advani J, and Rajagopalan P, et al (2018) Cigarette smoke and chewing tobacco alter expression of different sets of miRNAs in oral keratinocytes Sci Rep 8 https://doi.org/10.1038/s41598-018-25498-2
57. Arunkumar G, Deva Magendhra Rao AK, and Manikandan M, et al (2018) Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma Oncol Lett 15 649–657 https://doi.org/10.3892/ol.2017.7296 PMID: 29375721 PMCID: 5766066
58. Thomas AM, Gleber-Netto FO, and Fernandes GR, et al (2014) Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms BMC Microbiol 14 250 https://doi.org/10.1186/s12866-014-0250-2 PMID: 25278091 PMCID: 4186948
59. Vallès Y, Inman CK, and Peters BA, et al (2018) Types of tobacco consumption and the oral microbiome in the United Arab Emirates Healthy Future (UAEHFS) Pilot Study Sci Rep 8 https://doi.org/10.1038/s41598-018-29730-x PMID: 30054546 PMCID: 6063860
60. Wu J, Peters BA, and Dominianni C, et al (2016) Cigarette smoking and the oral microbiome in a large study of American adults ISME J 10 2435–2446 https://doi.org/10.1038/ismej.2016.37 PMID: 27015003 PMCID: 5030690
61. Grine G, Royer A, and Terrer E, et al (2019) Tobacco smoking affects the salivary gram-positive bacterial population Front Public Heal 7 196 https://doi.org/10.3389/fpubh.2019.00196
62. Beghini F, Renson A, and Zolnik CP, et al (2019) Tobacco exposure associated with oral microbiota oxygen utilization in the New York City Health and Nutrition Examination Study Ann Epidemiol 34 18–25.e3 https://doi.org/10.1016/j.annepidem.2019.03.005 PMID: 31076212 PMCID: 6548637
63. Mason MR, Preshaw PM, and Nagaraja HN, et al (2015) The subgingival microbiome of clinically healthy current and never smokers ISME J 9 268–272 https://doi.org/10.1038/ismej.2014.114
64. Shchipkova AY, Nagaraja HN, and Kumar PS (2010) Subgingival microbial profiles of smokers with periodontitis J Dent Res 89 1247–1253 https://doi.org/10.1177/0022034510377203 PMID: 20739702 PMCID: 3318026
65. Winkelhoff AJ van, Bosch-Tijhof CJ, and Winkel EG, et al (2001) Smoking affects the subgingival microflora in periodontitis J Periodontol 72 666–671 https://doi.org/10.1902/jop.2001.72.5.666 PMID: 11394403
66. Shakhatreh MAK, Khabour OF, and Alzoubi KH, et al (2018) Alterations in oral microbial flora induced by waterpipe tobacco smoking Int J Gen Med 11 47–54 https://doi.org/10.2147/IJGM.S150553 PMID: 29440924 PMCID: 5799848
67. Pushalkar S, Paul B, and Li Q, et al (2020) Electronic cigarette aerosol modulates the oral microbiome and increases risk of infection IScience 23(3) 100884 https://doi.org/10.1016/j.isci.2020.100884 PMID: 32105635 PMCID: 7113564
68. Liu M, Jin J, and Pan H, et al (2016) Effect of smokeless tobacco products on human oral bacteria growth and viability Anaerobe 42 152–161 https://doi.org/10.1016/j.anaerobe.2016.10.006 PMID: 27756619 PMCID: 5693344
69. Hernandez BY, Zhu X, and Goodman MT, et al (2017) Betel nut chewing, oral premalignant lesions, and the oral microbiome PLoS One 12(2) e0172196 https://doi.org/10.1371/journal.pone.0172196 PMID: 28225785 PMCID: 5321455
70. Tateda M, Shiga K, and Saijo S, et al (2000) Streptococcus anginosus in head and neck squamous cell carcinoma: implication in carcinogenesis Int J Mol Med 6 699–703 https://doi.org/10.3892/ijmm.6.6.699 PMID: 11078831
71. Sasaki M, Yamaura C, and Ohara-Nemoto Y, et al (2005) Streptococcus anginosus infection in oral cancer and its infection route Oral Dis 11 151–156 https://doi.org/10.1111/j.1601-0825.2005.01051.x PMID: 15888105
72. Morita E, Narikiyo M, and Yano A, et al (2003) Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer Cancer Sci 94 492–496 https://doi.org/10.1111/j.1349-7006.2003.tb01471.x PMID: 12824872
73. Vital M, Howe AC, and Tiedje JM (2014) Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data MBio 5 https://doi.org/10.1128/mBio.00889-14 PMID: 24757212 PMCID: 3994512
74. Miyazaki Y, Kikuchi K, and González-Alva P, et al (2010) Association of butyric acid produced by periodontopathic bacteria with progression of oral cancer J Cancer Sci Ther 2 26–32 https://doi.org/10.4172/1948-5956.1000019
75. Wu X, Wu Y, and He L, et al (2018) Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer J Cancer 9 2510–2517 https://doi.org/10.7150/jca.25324 PMID: 30026849 PMCID: 6036887
76. Bagaitkar J, Daep CA, and Patel CK, et al (2011) Tobacco smoke augments Porphyromonas gingivalis - Streptococcus gordonii biofilm formation PLoS One 6 27386 https://doi.org/10.1371/journal.pone.0027386
77. Bagaitkar J, Demuth DR, and Daep CA, et al (2010) Tobacco upregulates P. gingivalis fimbrial proteins which induce TLR2 hyposensitivity PLoS One 5 https://doi.org/10.1371/journal.pone.0009323 PMID: 20454607 PMCID: 2864253
78. Hanel AN, Herzog HM, and James MG, et al (2020) Effects of oral commensal streptococci on porphyromonas gingivalis invasion into oral epithelial cells Dent J 8 https://doi.org/10.3390/dj8020039
79. Olsen I and Yilmaz Ö (2019) Possible role of Porphyromonas gingivalis in orodigestive cancers J Oral Microbiol 11 https://doi.org/10.1080/20002297.2018.1563410
80. Katz J, Onate MD, and Pauley KM, et al (2011) Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma Int J Oral Sci 3 209–215 https://doi.org/10.4248/IJOS11075 PMID: 22010579 PMCID: 3469978
81. Wu J, Zheng M, and Zhang M, et al (2018) Porphyromonas gingivalis promotes 4-nitroquinoline-1-oxide-induced oral carcinogenesis with an alteration of fatty acid metabolism Front Microbiol 9 2081 https://doi.org/10.3389/fmicb.2018.02081 PMCID: 6131559
82. Dickinson BC, Moffatt CE, and Hagerty D, et al (2011) Interaction of oral bacteria with gingival epithelial cell multilayers Mol Oral Microbiol 26 210–220 https://doi.org/10.1111/j.2041-1014.2011.00609.x PMID: 21545698 PMCID: 3248246
83. Castillo G del V, Blanc SL de, and Sotomayor CE, et al (2018) Study of virulence factor of Candida species in oral lesions and its association with potentially malignant and malignant lesions Arch Oral Biol 91 35–41 https://doi.org/10.1016/j.archoralbio.2018.02.012
84. Bokor-Bratic M, Cankovic M, and Dragnic N (2013) Unstimulated whole salivary flow rate and anxiolytics intake are independently associated with oral Candida infection in patients with oral lichen planus Eur J Oral Sci 121 427–433 https://doi.org/10.1111/eos.12073 PMID: 24028590
85. Singh SK, Gupta A, and Rajan SY, et al (2014) Correlation of presence of Candida and epithelial dysplasia in oral mucosal lesions J Clin Diagnostic Res 8
86. Nagy KN, Sonkodi I, and Szöke I, et al (1998) The microflora associated with human oral carcinomas Oral Oncol 34 304–308 https://doi.org/10.1016/S1368-8375(98)80012-2 PMID: 9813727
87. Gall F, Colella G, and Di Onofrio V, et al (2013) Candida spp. in oral cancer and oral precancerous lesions New Microbiol 36 283–288 PMID: 23912870
88. Sheth CC, Makda K, and Dilmahomed Z, et al (2016) Alcohol and tobacco consumption affect the oral carriage of Candida albicans and mutans streptococci Lett Appl Microbiol 63 254–259 https://doi.org/10.1111/lam.12620 PMID: 27450704
89. Abduljabbar T, Hussain M, and Adnan T, et al (2017) Comparison of oral Candida species prevalence and carriage among gutka-chewers and betel-quid chewers J Pak Med Assoc 67 350–354 PMID: 28303980
90. Semlali A, Killer K, and Alanazi H, et al (2014) Cigarette smoke condensate increases C. albicans adhesion, growth, biofilm formation, and EAP1, HWP1 and SAP2 gene expression BMC Microbiol 14 61 https://doi.org/10.1186/1471-2180-14-61 PMID: 24618025 PMCID: 3995653
91. Baboni FB, Guariza Filho O, and Moreno AN, et al (2010) Influence of cigarette smoke condensate on cariogenic and candidal biofilm formation on orthodontic materials Am J Orthod Dentofac Orthop 138 427–434 https://doi.org/10.1016/j.ajodo.2009.05.023
92. Alanazi H, Semlali A, and Perraud L, et al (2014) Cigarette smoke-exposed candida albicans increased chitin production and modulated human fibroblast cell responses Biomed Res Int 2014 https://doi.org/10.1155/2014/963156 PMID: 25302312 PMCID: 4180399
93. Karkowska-Kuleta J, Rapala-Kozik M, and Kozik A (2009) Fungi pathogenic to humans: molecular bases of virulence of Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus.Acta Biochim Pol 56 211–224 https://doi.org/10.18388/abp.2009_2452
94. Patil S, Rao RS, and Raj AT (2017) Oral candidal carriage in subjects with pure vegetarian and mixed dietary habits J Clin Diagnostic Res 11
95. Al-Hebshi NN, Borgnakke WS, and Johnson NW (2019) The microbiome of oral squamous cell carcinomas: a functional perspective Curr Oral Heal Rep 6 145–160 https://doi.org/10.1007/s40496-019-0215-5
96. Hooper SJ, Crean SJ, and Fardy MJ, et al (2007) A molecular analysis of the bacteria present within oral squamous cell carcinoma J Med Microbiol 56 1651–1659 https://doi.org/10.1099/jmm.0.46918-0 PMID: 18033835
97. Zhao H, Chu M, and Huang Z, et al (2017) Variations in oral microbiota associated with oral cancer Sci Rep 7 1–10 https://doi.org/10.1038/s41598-017-11779-9
98. Zhang L, Liu Y, and Zheng HJ, et al (2020) The oral microbiota may have influence on oral cancer Front Cell Infect Microbiol 9 476 https://doi.org/10.3389/fcimb.2019.00476 PMID: 32010645 PMCID: 6974454
99. Perera M, Al-Hebshi NN, and Speicher DJ, et al (2016) Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio-pathogenic bacteria J Oral Microbiol 8 https://doi.org/10.3402/jom.v8.32762 PMID: 27677454 PMCID: 5039235
100. Bolz J, Dosá E, and Schubert J, et al (2014) Bacterial colonization of microbial biofilms in oral squamous cell carcinoma Clin Oral Investig 18 409–414 https://doi.org/10.1007/s00784-013-1007-2
101. Nagy K, Szöke I, and Sonkodi I, et al (2000) Inhibition of microflora associated with oral malignancy Oral Oncol 36 32–36 https://doi.org/10.1016/S1368-8375(99)00046-9 PMID: 10889916
102. Pushalkar S, Ji X, and Li Y, et al (2012) Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma BMC Microbiol 12 144 https://doi.org/10.1186/1471-2180-12-144 PMID: 22817758 PMCID: 3507910
103. Lee WH, Chen HM, and Yang SF, et al (2017) Bacterial alterations in salivary microbiota and their association in oral cancer Sci Rep 7 https://doi.org/10.1038/s41598-017-16418-x
104. Bollati V, Fabris S, and Pegoraro V, et al (2017) Possible mycological etiology of oral mucosal cancer: catalytic potential of infecting Candida aibicans and other yeasts in production of N -nitrosobenzylmethylamine Carcinogenesis 30 1330–1335 https://doi.org/10.1093/carcin/bgp149
105. Hooper SJ, Wilson MJ, and Crean SJ (2009) Exploring the link between microorganisms and oral cancer: a systematic review of the literature Head Neck 31 1228–1239 https://doi.org/10.1002/hed.21140 PMID: 19475550
106. Shukla K, Vun I, and Lov I, et al (2019) Role of Candida infection in the malignant transformation of oral leukoplakia: A systematic review of observational studies Transl Res Oral Oncol 4 2057178X1982822 https://doi.org/10.1177/2057178X19828229
107. Sankari SL, Gayathri K, and Balachander N, et al (2015) Candida in potentially malignant oral disorders J Pharm Bioallied Sci 7 S162–S164 https://doi.org/10.4103/0975-7406.155886 PMID: 26015698 PMCID: 4439658
108. Hooper SJ, Crean SJ, and Lewis MAO, et al (2006) Viable bacteria present within oral squamous cell carcinoma tissue J Clin Microbiol 44 1719–1725 https://doi.org/10.1128/JCM.44.5.1719-1725.2006 PMID: 16672398 PMCID: 1479175
109. Vivarelli S, Salemi R, and Candido S, et al (2019) Gut microbiota and cancer: from pathogenesis to therapy Cancers (Basel) 11 https://doi.org/10.3390/cancers11010038
110. García-Castillo V, Sanhueza E, and McNerney E, et al (2016) Microbiota dysbiosis: A new piece in the understanding of the carcinogenesis puzzle J Med Microbiol 65 1347–1362 https://doi.org/10.1099/jmm.0.000371 PMID: 27902422
111. Sheflin AM, Whitney AK, and Weir TL (2014) Cancer-promoting effects of microbial dysbiosis Curr Oncol Rep 16 1–9 https://doi.org/10.1007/s11912-014-0406-0
112. Kosumi K, Mima K, and Baba H, et al (2018) Dysbiosis of the gut microbiota and colorectal cancer: the key target of molecular pathological epidemiology J Lab Precis Med 3 76 https://doi.org/10.21037/jlpm.2018.09.05 PMID: 30345420 PMCID: 6195365
113. Meurman JH (2010) Oral microbiota and cancer J Oral Microbiol 2 https://doi.org/10.3402/jom.v2i0.5195 PMID: 21523227 PMCID: 3084564
114. la Rosa GRM, Gattuso G, and Pedullà E, et al (2020) Association of oral dysbiosis with oral cancer development (Review) Oncol Lett 19 3045–3058 https://doi.org/10.3892/ol.2020.11441 PMID: 32211076 PMCID: 7079586
115. Lax AJ (2005) Bacterial toxins and cancer - a case to answer? Nat Rev Microbiol 3 343–349 https://doi.org/10.1038/nrmicro1130 PMID: 15806096
116. Zhang G and Ghosh S (2004) Molecular mechanisms of NF-κB activation induced by bacterial lipopolysaccharide through Toll-like receptors J Endotoxin Res 6 453–457 https://doi.org/10.1179/096805100101532414
117. Coussens LM and Werb Z (2002) Inflammation and cancer Nature 420 860–867 https://doi.org/10.1038/nature01322 PMID: 12490959 PMCID: 2803035
118. Philip M, Rowley DA, and Schreiber H (2004) Inflammation as a tumor promoter in cancer induction Semin Cancer Biol 14 433–439 https://doi.org/10.1016/j.semcancer.2004.06.006 PMID: 15489136
119. Lin WW and Karin M (2007) A cytokine-mediated link between innate immunity, inflammation, and cancer J Clin Invest 117 1175–1183 https://doi.org/10.1172/JCI31537 PMID: 17476347 PMCID: 1857251
120. Harrandah A, Chukkapalli SS, and Dunn WJ, et al (2019) Abstract 2824: Modulation of oral cancer cells survival and invasiveness by fusobacteria Cancer Res 79 2824 https://doi.org/10.1158/1538-7445.am2019-2824
121. Ha NH, Woo BH, and Kim DJ, et al (2015) Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties Tumor Biol 36 9947–9960 https://doi.org/10.1007/s13277-015-3764-9
122. Yilmaz Ö, Jungas T, and Verbeke P, et al (2004) Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis Infect Immun 72 3743–3751 https://doi.org/10.1128/IAI.72.7.3743-3751.2004 PMID: 15213114 PMCID: 427421
123. Mao S, Park Y, and Hasegawa Y, et al (2007) Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis Cell Microbiol 9 1997–2007 https://doi.org/10.1111/j.1462-5822.2007.00931.x PMID: 17419719 PMCID: 2886729
124. Yao L, Jermanus C, and Barbetta B, et al (2010) Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells Mol Oral Microbiol 25 89–101 https://doi.org/10.1111/j.2041-1014.2010.00569.x PMID: 20331797 PMCID: 2894563
125. Kuboniwa M, Hasegawa Y, and Mao S, et al P. gingivalis accelerates gingival epithelial cell progression through the cell cycle Microbes Infect 10 122–128 https://doi.org/10.1016/j.micinf.2007.10.011 PMID: 18280195 PMCID: 2311419
126. Inaba H, Sugita H, and Kuboniwa M, et al (2014) Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation Cell Microbiol 16 131–145 https://doi.org/10.1111/cmi.12211 PMCID: 3939075
127. Pan C, Xu X, Tan L, Lin L, Pan Y. The effects of Porphyromonas gingivalis on the cell cycle progression of human gingival epithelial cells. Oral Dis 2014;20:100–8. https://doi.org/10.1111/odi.12081
128. Uitto VJ, Baillie D, and Wu Q, et al (2005) Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells Infect Immun 73 1171–1179 https://doi.org/10.1128/IAI.73.2.1171-1179.2005 PMID: 15664960 PMCID: 547012
129. Zhou Y, Sztukowska M, and Wang Q, et al (2015) Noncanonical activation of β-catenin by Porphyromonas gingivalis Infect Immun 83 3195–3203 https://doi.org/10.1128/IAI.00302-15
130. Zhang W-l, Wang S-s, and Wang H-f, et al (2019) Who is who in oral cancer? Exp Cell Res 384 111634 https://doi.org/10.1016/j.yexcr.2019.111634 PMID: 31541617
131. Gallimidi AB, Fischman S, and Revach B, et al (2015) Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model Oncotarget 6 22613–22623 https://doi.org/10.18632/oncotarget.4209 PMCID: 4673186
132. Zakikhany K, Naglik JR, and Schmidt-westhausen A, et al (2007) In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination Cell Microbiol 9 2938–2954 https://doi.org/10.1111/j.1462-5822.2007.01009.x PMID: 17645752
133. Naglik JR, König A, and Hube B, et al (2017) Candida albicans–epithelial interactions and induction of mucosal innate immunity Curr Opin Microbiol 40 104–112 https://doi.org/10.1016/j.mib.2017.10.030 PMID: 29156234 PMCID: 5733685
134. Phan QT, Myers CL, and Fu Y, et al (2007) Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells PLoS Biol 5 0543–0557 https://doi.org/10.1371/journal.pbio.0050064
135. Martin R, Wächtler B, and Schaller M, et al (2011) Host-pathogen interactions and virulence-associated genes during Candida albicans oral infections Int J Med Microbiol 301 417–422 https://doi.org/10.1016/j.ijmm.2011.04.009 PMID: 21555244
136. Phan QT, Fratti RA, and Prasadarao NV, et al (2005) N-cadherin mediates endocytosis of Candida albicans by endothelial cells J Biol Chem 280 10455–10461 https://doi.org/10.1074/jbc.M412592200 PMID: 15632157
137. Wächtler B, Wilson D, and Haedicke K, et al (2011) From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells PLoS One 6 https://doi.org/10.1371/journal.pone.0017046 PMID: 21407800 PMCID: 3044159
138. Moyes DL, Wilson D, and Richardson JP, et al (2016) Candidalysin is a fungal peptide toxin critical for mucosal infection Nature 532 64–68 https://doi.org/10.1038/nature17625 PMID: 27027296 PMCID: 4851236
139. Engku Nasrullah Satiman EAF, Ahmad H, and Ramzi AB, et al (2020) The role of Candida albicans candidalysin ECE1 gene in oral carcinogenesis J Oral Pathol Med jop.13014 https://doi.org/10.1111/jop.13014
140. Krogh P (1990) The role of yeasts in oral cancer by means of endogenous nitrosation Acta Odontol Scand 48 85–88 https://doi.org/10.3109/00016359009012738 PMID: 2181815
141. Sanjaya PR, Gokul S, and Gururaj Patil B, et al (2011) Candida in oral pre-cancer and oral cancer Med Hypotheses 77 1125–1128 https://doi.org/10.1016/j.mehy.2011.09.018 PMID: 21978967
142. O’grady JF and Reade PC (1992) Candida albicans as a promoter of oral mucosal neoplasia Carcinogenesis 13 783–786 [https://doi.org/10.1093/carcin/13.5.783] https://doi.org/10.1093/carcin/13.5.783
143. Gümüa ZH, Du B, and Kacker A, et al (2008) Effects of tobacco smoke on gene expression and cellular pathways in a cellular model of oral leukoplakia Cancer Prev Res 1 100–111 https://doi.org/10.1158/1940-6207.CAPR-08-0007
144. Sasaki M, Ohara-Nemoto Y, and Tajika S, et al (2001) Antigenic characterisation of a novel Streptococcus anginosus antigen that induces nitric oxide synthesis by murine peritoneal exudate cells J Med Microbiol 50 952–958 https://doi.org/10.1099/0022-1317-50-11-952 PMID: 11699591
145. Patil S, Arakeri G, and Alamir AWH, et al (2020) Is toombak a risk factor for oral leukoplakia and oral squamous cell carcinoma? A systematic review J Oral Pathol Med 49 103–109 https://doi.org/10.1111/jop.12954
146. Negi M, Sepolia N, and Panwar S, et al (2019) Prevalence of oral parameters in smokeless tobacco-associated precancer J Fam Med Prim Care 8 3956 https://doi.org/10.4103/jfmpc.jfmpc_667_19
147. Hu X, Zhang Q, and Hua H, et al (2016) Changes in the salivary microbiota of oral leukoplakia and oral cancer Oral Oncol 56 e6–e8 https://doi.org/10.1016/j.oraloncology.2016.03.007 PMID: 27026576
148. Bakaris S, Okur E, and Yildirim I, et al (2007) Ki-67 protein expression in smokeless tobacco (Maras powder)-induced oral mucosal lesions Toxicol Mech Methods 17 567–574 https://doi.org/10.1080/15376510701523823 PMID: 20020883
149. Rajagopalan P, Patel K, and Jain AP, et al (2018) Molecular alterations associated with chronic exposure to cigarette smoke and chewing tobacco in normal oral keratinocytes Cancer Biol Ther 19 773–785 https://doi.org/10.1080/15384047.2018.1470724 PMID: 29723088 PMCID: 6154853
150. Schmidt BL, Kuczynski J, and Bhattacharya A, et al (2014) Changes in abundance of oral microbiota associated with oral cancer PLoS One 9 e98741 https://doi.org/10.1371/journal.pone.0098741 PMID: 24887397 PMCID: 4041887
151. Rohatgi N, Matta A, and Kaur J, et al (2006) Novel molecular targets of smokeless tobacco (khaini) in cell culture from oral hyperplasia Toxicology 224 1–13 https://doi.org/10.1016/j.tox.2006.03.014 PMID: 16730401