Assessment of mismatch repair proteins (MLH1 and MSH2) and p53 immunohistochemical expression in prostatic carcinoma: association with different clinicopathologic characteristics
Hend S Abo Safia1,2,a, Ahmed F Ghaith3,b, Eman E Farghal4,c and Basma S Amer1,d
1Pathology Department, Faculty of Medicine, Tanta University, El Geesh Street, Tanta 31527, Egypt
2Department of Basic Medical Sciences, Faculty of Medicine, Ibn Sina University for Medical Sciences, Amman 16197, Jordan
3Department of Urology, Tanta University, El Geesh Street, Tanta 31527 Egypt
4Department of Clinical and Chemical Pathology, Faculty of Medicine, Tanta University, El Geesh Street, Tanta 31527, Egypt
ahttp://orcid.org/0000-0001-9117-4441
bhttp://orchid.org/0000-0002-9102-1821
chttp://orcid.org/0000-0002-7562-055X
dhttp://orchid.org/0000-0003-0123-3255
Abstract
Background: Prostate cancer (PCa) is one of the most heritable human cancers and it is the second most frequent malignancy in men worldwide. It accounts for a significant morbidity and mortality throughout the world. PCa with mismatch repair (MMR) deficiency often has aggressive clinical and histological features, but its rarity prevents the analysis of the underlying biology. Therefore, in this study, we aimed to evaluate the immunohistochemical expression of MMR proteins and P53 in PCa.
Materials and methods: Fifty cases of PCa were histologically examined. The MMR proteins and P53 immunoexpression were assessed. Also, P53 serum concentration levels using ELIZA was measured and pre-operative prostatic specific antigen (PSA) serum levels were obtained.
Results: There was a significant positive relation between mutS homologue 2 (MSH2) immunoexpression and both PSA serum level and P53 serum concentration (p value 0.001*). Also, there was a significant relation between MSH2 immunoexpression and tumour size, nodal metastasis, distant metastasis and grade grouping. While mutL homologue 1 (MLH1) immunoexpression showed a significant relation with human P53 serum concentrations only (p value 0.035*). Moreover, MLH1 immunoexpression showed only significant relation with nodal metastasis and tumour burden, p value was 0.033* and 0.001*, respectively.
Conclusion: MMR protein loss, especially MSH2, was seen in a significant subset of PCa. Interestingly, it was associated with significantly higher levels of serum PSA and p53. Moreover, it may be associated with unfortunate prognostic features as large tumour size, higher grade grouping and finally nodal and distant metastasis.
Keywords: prostate cancer, mismatch repair proteins MSH2, MLH1, P53
Correspondence to: Hend S Abo Safia
Email: hend.abosafeia@med.tanta.edu.eg
Published: 04/09/2025
Received: 11/02/2025
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Background
Prostate cancer (PCa) is considered the second most common cancer and the fifth leading cause of cancer mortality among aging men worldwide. There is an urgent need for clinically useful biomarkers for early detection of PCa to lessen the liabilities of overtreatment and accompanying tumour morbidity [25].
There are many risk factors for the development of PCa, including age, ethnicity and genetic factors. PCa is considered one of the most heritable human cancers and its heritability was estimated to be 57% in some populations, which is higher than the rates for almost all other cancers, including breast, kidney and ovarian cancer (31%, 38% and 39%, respectively) [17].
The typical morphology of prostate carcinoma is 'small gland' which has rigid round lumens, and often has characteristic infiltrative growth pattern. But the 'large gland' morphology is not infrequently detected [16]. They are large, crowded with irregular contour, papillary inholding and luminal undulation. Large gland prostate cancer may resemble adjacent benign glands architecturally, but are often larger than the latter [34]. Also, malignant glands are lined with cytologically atypical cells with nucleomegaly and prominent nucleoli [5].
The modifications of the International Society of Urological Pathology (ISUP) consensus conference in 2014 in Chicago were incorporated into the 2022 World Health Organization (WHO) Classification of Tumours of the Urinary System and Male Genital Organs. In the past 5 years, further new data in the Gleason pattern quantities, tumour growth patterns and clinical practice advancements such as widespread introduction of multiparametric magnetic resonance imaging-targeted biopsies or fusion ultrasound/magnetic resonance imaging biopsies have added to challenges in reporting and grading for pathologists [30].
The mismatch repair (MMR) deficiency causes functional abnormality of MMR proteins, resulting in their loss on immunohistochemistry (IHC). MMR is an excision-re synthesis system which acts as a sensor for DNA damage, correcting the mismatches generated during DNA replication. So, their deficiency leads to the accumulation of errors during DNA replication, especially in repetitive sequences known as microsatellites [19]. Defects in DNA MMR proteins are considered permissive for carcinogenesis, giving rise to microsatellite instability (MSI) and their role has been extensively studied in colorectal and endometrial cancer [1].
The recent studies on MMR protein expression in prostate tumours revealed that approximately 10% of advanced/metastatic prostate tumours have an underlying somatic and/or germline inactivation of genes in the MMR family (mutS homologue 2 (MSH2), MSH6, mutL homologue 1 (MLH1) or PMS2) [13,15]. This is similar to what has been detected in colorectal carcinoma [18, 24].
Many studies have searched for MMR defects in advanced PCa, but the relative frequency and clinical significance of MMR alterations in early PCa are less certain [6, 12].
As in many human cancers, p53 tumour suppressor gene mutations are a frequent genetic event in PCa, and can be detected in up to 94% of cases. With the exception of Li-Fraumeni syndrome and a very little fraction of other neoplasms in high-risk groups, most of the p53 abnormalities represent late somatic alterations in the process of carcinogenesis [3].
Many previous studies have focused on the role of p53 in prostatic cancer but most of them claimed that p53 gene mutations are not frequently detected in early-stage prostatic cancer. Recently, increasing evidence suggests that loss of p53 function may be an important early step in disease progression. Moreover, clonally altered p53 in earlier stage disease may be a prognostic marker. In addition, several recent studies have shown frequent p53 alterations in hormone-refractory prostatic cancer [14, 29].
In this study, we evaluated the relationship between the immunohistochemical expression of MMR proteins MLH1, MSH2 and p53 in prostatic cancer and their relation to the available clinicopathological features.
Methods
The current retrospective study included 50 cases diagnosed as prostatic cancer during the period from February 2021 to October 2023. The study was approved by Research Ethics Committee, Faculty of Medicine, Tanta University, with approval number (36150/12/22). Written informed consent forms were collected once all study participants received information about it. This retrospective study was conducted at the Urology together with Pathology and Clinical Pathology Departments of Tanta University Hospital. Serum levels of prostatic specific antigen (PSA) and P53 levels were also assessed.
All patients underwent digital rectal examination and 12 cores transrectal ultrasound-guided prostatic biopsy, which was done by an expert urologist. The patients diagnosed as prostatic adenocarcinoma were subjected to computerised tomography for abdomen and pelvis and bone scintigraphy for clinical staging.
All cases were subjected to routine haematoxylin and eosin staining and examined by two pathologists to confirm the diagnosis.
Blood sampling
Determination of p53 and PSA parameters
For the patients who were diagnosed as prostatic adenocarcinoma, blood samples were collected under strict aseptic conditions into serum vacutainer tubes and approximately after 30 minutes tubes were centrifuged at 6,000 revolutions per minute for 10 minutes at room temperature to obtain serum for p53 and PSA analysis. Serum samples were stored at −80°C until analysis of the dependent variables. P53 and PSA concentrations were measured spectrophotometrically using a plate reader (Multiskan Spectrum, Thermo Lab Systems). A p53 enzyme-linked immunosorbent assay (ELISA) kit (Human p53 ELISA, Catalog No: BMS256; Bender MedSystems GmbH, Austria, Europe) was used to measure p53 protein concentration in serum samples according to the manufacturer’s protocol. Serum PSA was measured using ELISA kit (PSA ELISA Catalog Number SE120106) according to the manufacturer’s protocol. To eliminate inter-assay variance, all samples for a particular assay were thawed once and analysed in the same assay run.
Histopathological evaluation
Sections from the studied cases were subjected to hematoxylin and eosin staining. Gleason score was assigned according to the 2022 WHO classification ISUP consensus ISUP [32].
Immunohistochemical staining
Tumour sections (of 5 μm thickness) on positively charged slides were left to dry for 30 minutes at 37°C. Sections were then deparaffinised and antigen retrieval was performed in Dako PT link unit using high and low PH EnVision FLEX antigen retrieval solutions (reaching 97°C for 20 minutes). Immunostaining was achieved in Dako Autostainer Link 48. Briefly, peroxidase blocking reagent was applied, followed by incubation with primary antibodies for 30 minutes. After that horseradish peroxidase polymer was applied for 20 minutes and diaminobenzidine was applied as chromogen. The slides were then counterstained by hematoxylin.
The primary antibodies applied in this study were:
1- MLH1, a monoclonal antibody (mouse anti-hMLH1 antibody (clone ES05, 1:50; Dako, Glostrup, Denmark),
2- MSH2, a monoclonal mouse anti-hMSH2 antibody (clone FE11,1:50 Dako Glostrup, Denmark),
3- P53, a monoclonal antibody, clone DO-7; 1:50 Dako, USA).
Interpretation immunohistochemical stains
Interpretation of MLH1, MSH2 immunostaining results
If the tumour showed total absence of nuclear staining with the adjacent normal tissue showing normal nuclear staining, the case was regarded as deficient MMR. While tumours with normal nuclear staining in the neoplastic cells were regarded as having proficient MMR [8, 9].
Interpretation of P53 immunostaining results
Immunoreactivity was regarded as positive when brown staining was localised to the nucleus of the neoplastic cells. The evaluation of the IHC slides was done semi-quantitatively, and the staining was scored based on intensity as follows: 0, negative staining; 1+, weak staining; 2+, moderate staining and 3+, strong staining. For the statistical analysis, the cases were categorised in two groups, 0 and 1+ as low expression group and +2 and +3 as high expression group. The adjacent benign glands should not show more than weak, partial staining, if any. Negative staining pertains to no staining or focal, weak fine granular staining [21].
Positive controls for p53 nuclear accumulation used with each run of staining were a colon cancer specimen with p53 missense mutation.
Statistical analysis
Data were statistically analysed using Statistical Package for the Social Sciences (version 23.0). Data were expressed as frequencies for categorial data, while mean ± SD was used to represent numerical data. Chi square test was used to compare categorial data, while Fischer exact or Monte-Carlo tests were used when appropriate. Student t-test was used to compare numerical data. p values of less than 0.05 were considered statistically significant.
Results
Clinicopathological features
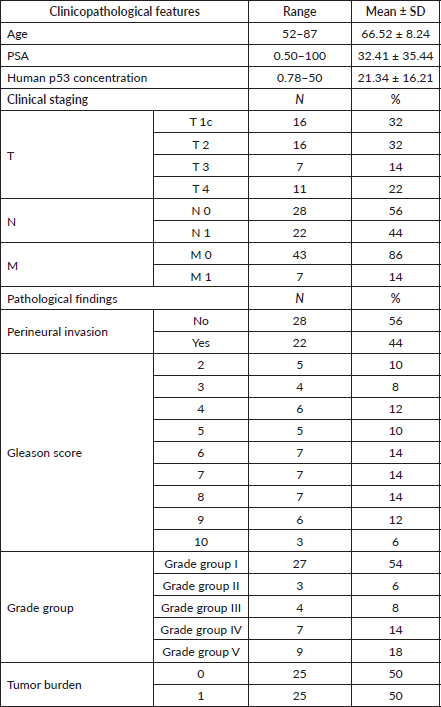
The present study included 50 cases of prostatic adenocarcinoma. Patient characteristics were shown in Table 1. The age of the patients ranged from 52 to 87 years old. PSA plasma levels ranged from 0.50 to 100 ng/dL. Human p53 serum concentration levels ranged from 0.78 to 50 U/mL.
Clinical staging
According to Table 1, 64% of cases were T1c and T2 equally, while 22% of cases were T4 and only 14% of cases were T3. Lymph node (LN) metastasis was found in 44% of cases. Usually, LNs with a short axis >8 mm in the pelvis and >10 mm outside the pelvis are considered malignant. Distant metastasis was only present in 14% of cases at the time of diagnosis.
Pathological findings
Regarding perineural invasion, it was detected in 44% of cases. Gleason score was defined and most of the studied cases (40%) were well differentiated prostatic adenocarcinoma, while moderately differentiated PCa and poorly differentiated PCa represented in 28% and 32% of studied cases, respectively, as shown in Table 1.
After careful histopathological examination (Figure 1) of all cases and defining the exact Gleason score for each case, Grade Grouping was done and 54% of cases were Grade Group 1, while Grade Group 2 was the least represented in the current study, 6% of cases. Grade Groups IV and V represented 14% and 18%, respectively.
Table 1. Clinicopathological features of studied cases.
MMR proteins immunoexpression results
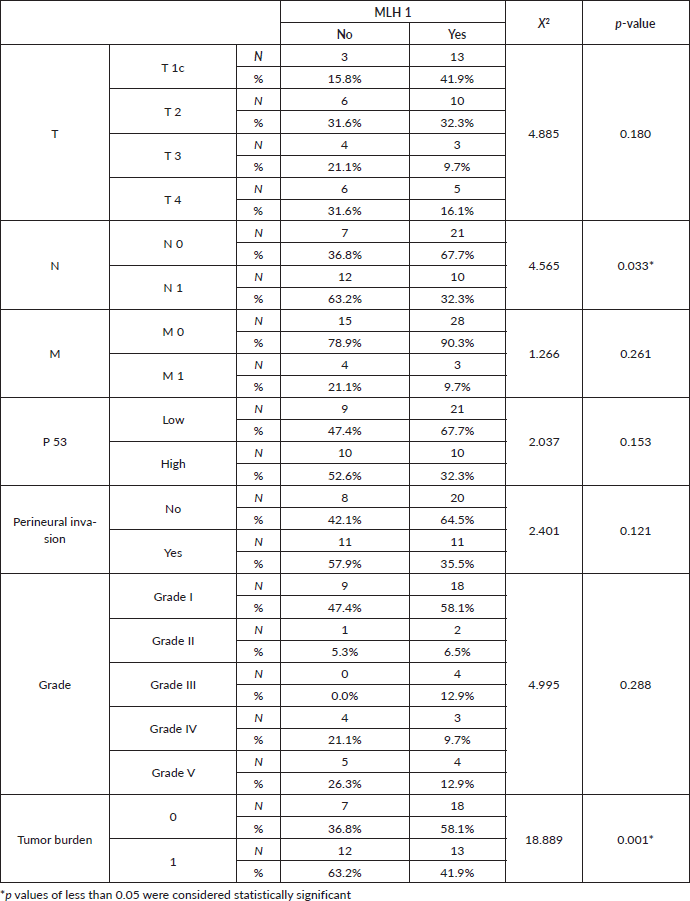
At the current study, MSH2 and MLH1 immunoexpression was absent in 36% and 38% of cases, respectively, as shown in Table 2 and Figures 2 and 3. Regarding the relation between each marker and the studied clinicopathological features and histopathological results, it is demonstrated in Tables 3–6. There was a significant positive relation between MSH2 immunoexpression and both PSA serum level and P53 serum concentration (p value 0.001*). Also, there was a significant relation between MSH2 immunoexpression and tumour size, nodal metastasis, distant metastasis and grade grouping. While MLH1 immunoexpression showed a significant relation with human P53 serum concentrations only (p value 0.035*), as shown in Table 5. Moreover, MLH1 immunoexpression showed only significant relation with nodal metastasis and tumour burden, p value was 0.033* and 0.001*, respectively.
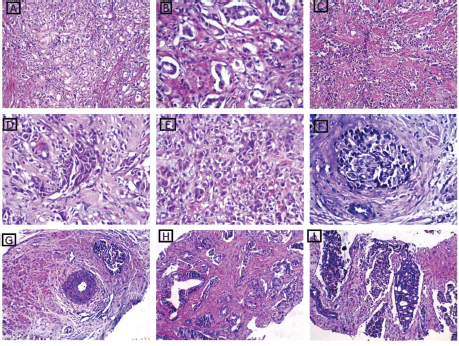
Figure 1. H&E-stained sections from studied PC cases. (a and b): A case of well-differentiated PC Gleason score 3 showing uniform glands with spaces between them. The glands are lined by a single layer of malignant cells (×200 and ×400, respectively. (c–e): Cases of poorly differentiated PC Gleason score 9 showing infiltrating fused glands admixed with sheets and cords of malignant cells ×200 and ×400, respectively. (f): A case of moderately differentiated PC Gleason score 7 showing perineural infiltration ×400. (g): a case of moderately differentiated PC Gleason score 7 showing malignant glands near muscular blood vessel ×400. (h and i): Moderately differentiated PC Gleason score 7 showing medium sized glands with undulating luminal contours and large branching glands with areas of cribriforming ×200.
Table 2. MMR proteins immunoexpression results.
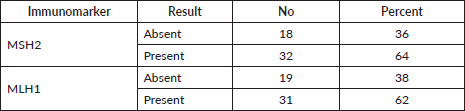
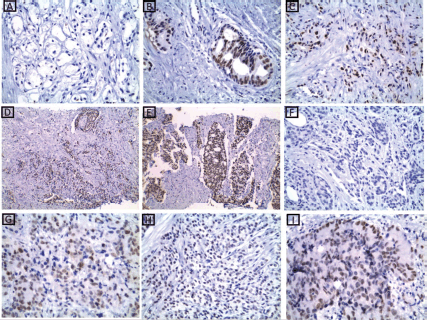
Figure 2. MSH2 immunohistochemical results. (a): A case of well-differentiated PC Gleason score 3 showing absent nuclear expression of MSH2 ×200. (b and c): Two cases of moderately differentiated PC Gleason score 7 showing retained nuclear expression of MSH2 ×400. (d): A case of poorly differentiated PC Gleason score 9 showing retained nuclear expression of MSH2 ×200. (e): A case of moderately differentiated PC Gleason score 7 showing retained nuclear expression of MSH2 ×200. (f): A case of moderately differentiated PC Gleason score 7 showing absent nuclear expression of MSH2 ×400. (g–i): cases of poorly differentiated PC Gleason score 9 and 10 showing retained nuclear expression of MSH2 ×400.
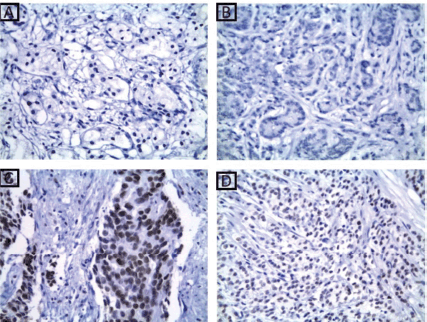
Figure 3. MLH1 immunohistochemical results. (a): A case of well-differentiated PC Gleason score 3 showing absent nuclear expression of MLH1 ×200. (b): A case of moderately differentiated PC Gleason score 7 showing absent nuclear expression of MLH1 ×400. (c): A case of moderately differentiated PC Gleason score 7 showing retained nuclear expression of MLH1 ×400. (d): A case of poorly differentiated PC Gleason score 9 showing retained nuclear expression of MLH1 ×400.
Table 3. Relation between MSH2 immunoexpression and clinicopathological features.
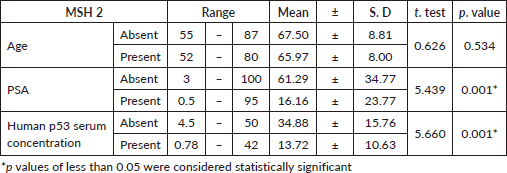
Table 4. Relation between MSH2 immunoexpression, clinical staging and histopathological features.
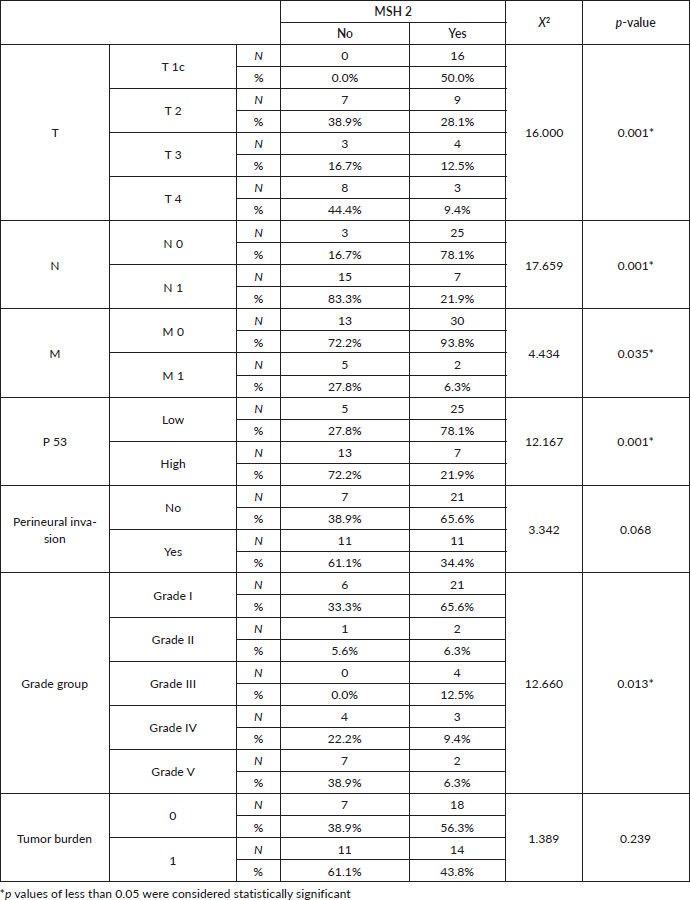
Table 5. Relation between MLH1 immunoexpression and clinicopathological features.
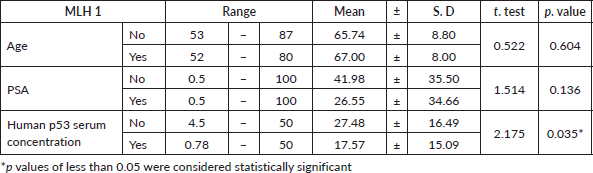
Table 6. Relation between MLH1 immunoexpression, clinical staging and histopathological features.
P53 protein immunoexpression results
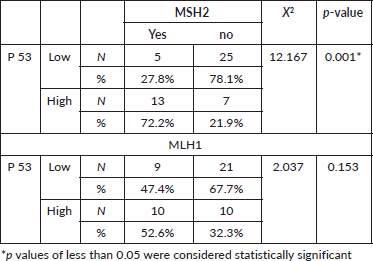
Tumour P53 expression was found to be low in 60% of studied prostatic carcinoma (PC) cases as shown in Table 7 and Figure 4. Regarding the relation between P53 immunoexpression and the available clinicopathological features, there was a significant positive relation between its expression and serum PSA levels and serum Human P53 concentrations (p value 0.001*) for both, as illustrated in Table 8. There was no statistically significant relation between P53 immunoexpression and any of the studied histopathological results as shown in Table 9.
Table 7. P53 immunoexpression in studied cases.
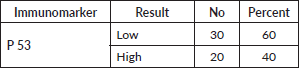
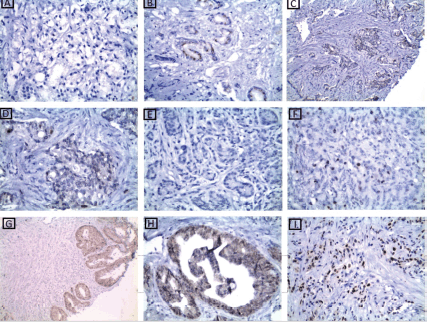
Figure 4. p53 immunohistochemical results. (a): A case of well-differentiated PC Gleason score 3 showing low nuclear expression of p53 ×200. (b): A case of moderately differentiated PC Gleason score 7 showing low nuclear expression of p53 ×400. (c and d): A case of moderately differentiated PC Gleason score 7 showing low nuclear expression of p53 ×200 and ×400, respectively. (e): A case of moderately differentiated PC Gleason score 7 showing low nuclear expression of p53 ×400. (f): A case of poorly differentiated PC Gleason score 9 showing low nuclear expression of p53 ×400. (g and h): A case of moderately differentiated PC Gleason score 7 showing high nuclear expression of p53 ×200 ×400, respectively. (i): A case of poorly differentiated PC Gleason score 9 showing high nuclear expression of p53 ×400.
Table 8. Relation between P53 immunoexpression and clinicopathological features.
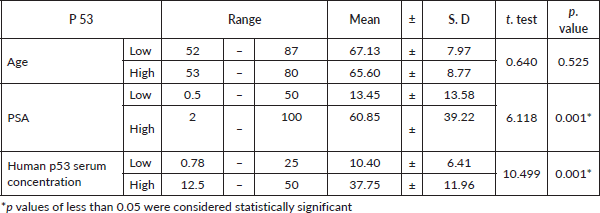
Table 9. Relation between P53 immunoexpression, clinical staging and histopathological results.
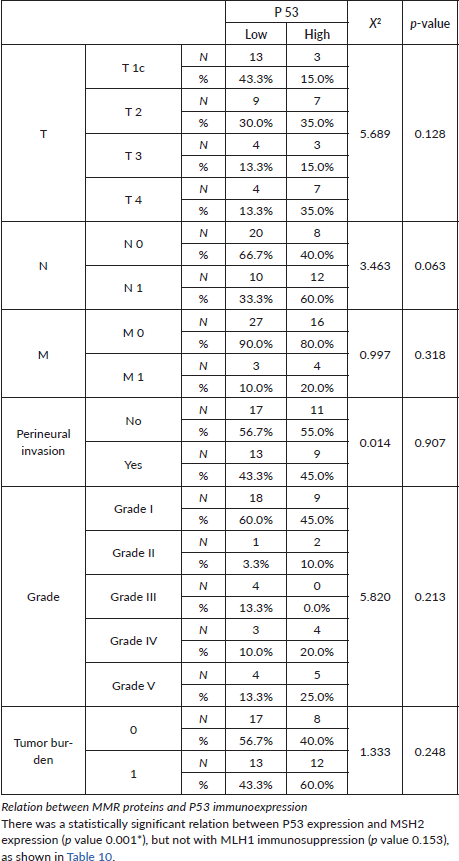
Table 10. Relation between P53 expression and MMR proteins immunoexpression.
Discussion
PCa ranks as the second most prevalent cancer among men and represents the fifth leading cause of cancer-related deaths worldwide. The wide spectrum of biological behaviour exhibited by prostatic neoplasms poses a difficult problem in predicting the clinical course for the individual patient. Histopathology together with the specified pathological markers plays a very important role in its preoperative and postoperative evaluation. Although PCa are slowly growing tumours, they vary widely in their aggressiveness [2].
Many traditional prognostic factors are present, such as tumour grade, clinical stage and pre-treatment PSA plasma levels, but they are of limited prognostic value for individual patient. So, the need for new prognostic molecular factors is necessary [11].
In recent years, some cases with PC have been linked with defects in MMR proteins. Worldwide, the detection of MMR proteins by IHC is routinely done in colorectal and endometrial adenocarcinoma. In a study by Hashemi et al [35] and another by Qasim et al [20], a high frequency of MMR deficiency in colorectal and endometrial adenocarcinoma was observed, respectively [20], but it is quite rare in PCa and its histology and molecular morphology were incompletely described. Thus, this study is trying to identify the features of PC with MSI [33].
In a study by Rodrigues et al [23], they revealed that impairment of MMR genes has been linked to the risk of PCa in men with Lynch syndrome. Moreover, it has been recently studied in sporadic cases of PCa and it has indeed been associated with worse patient outcomes, while favourable response to immune checkpoint blockade in those with MMR deficiency has also been reported. However, the clinical impact of MMR deficiency, especially that detected by IHC, in patients with PCa remains largely unknown [24].
In the current study, IHC expression of MSH2 and MLH1 was evaluated, in terms of their deficiency in prostatic adenocarcinoma, as they are known to play an important role in carcinogenesis, via MSI, particularly defects within the MSH2 gene. So, it provides valuable insights into the genetic characteristics of these tumours and help us identifying patients who may benefit from immunotherapy. Also, analysis of their relationship with respect to age, Gleason score, tumour size, nodal metastasis, tumour burden, grade group, PSA and P53 levels were done to assess their predictive role in predicting aggressive behaviour of PCa.
At the present study, MSH2 and MLH1 immunoexpression were absent in 36% and 38% of cases, respectively. Also, there was a significant positive relation between MSH2 immunoexpression and both PSA serum level and p53 serum concentrations. Also, there was a significant relation between MSH2 immunoexpression and tumour size, nodal metastasis, distant metastasis and grade grouping but MSH1 immunoexpression showed significant relations with only P53 serum concentration, tumour metastasis and tumour burden. Loss of at least one of the
MMR proteins was identified in 50 (22.7%) cases. These findings indicate a strong association between the deficiency of MMR proteins and biological and aggressive behaviours of invasive PCa.
A few immunohistochemical studies have reported the incidence of MMR deficiency in PCa. Sharma et al [26]reported that MLH1, MSH2, MSH6 and PMS2 were lost in 2 (0.9%), 6 (2.7%), 37 (16.8%) and 27 (12.3%) of PCa, respectively. They explained these low incidences by some limitations as using TMA consisting of only 1-mm tissue cores that may not be representative of the invasive cancer in each case, which may thus produce false-negative results.
According to Sharma et al [26], there were no statistically significant associations between MMR deficiency and patient age, family history of PCa, Gleason score or pT/pN stage. However, the levels of preoperative PSA were significantly (p  =  0.015) higher in patients with MMR deficiency (mean ±  SD: 9.12  ±  9.01  ng/mL) than in those without abnormal MMR (5.76  ±â€Š 3.17  ng/mL).
Among other studies, Guedes et al [8, 9] demonstrated that MSH2 loss was significantly more often seen in tumours with Gleason score 9–10/Grade Group 5 than in those with Gleason score ≤8/Grade Group ≤4. In addition, Wilczak et al [31] noted elevated expression of MLH1, MSH6 and PMS2 in PCa, which was associated with higher Gleason score or pT stage, LN metastasis or earlier biochemical recurrence. They concluded that high expression of MMR genes was associated with signs of genomic instability. So, as the number of deletions increased, the proportion of cancers showing strong MMR gene expression increased.
In accordance with previous observations and results, our research showed concurrent loss of MLH1 and MSH2. Moreover, the levels of preoperative PSA in patients with MMR deficiency were significantly higher than those without MMR deficiency. These results were recently confirmed by a study conducted by Grindedal et al [7] who found statistically significant increased incidence of prostatic cancer among MSH2 and MSH6 carriers, where the standardized incidence ratio was as high as 13 and 13.74, respectively, which also showed aggressive behaviour. Moreover, they concluded that their findings support continued PSA screening of MSH2 and MSH6 carriers.
TP53 inactivation is the genomic biomarker most consistently associated with adverse outcomes in primary and metastatic PCa. Many of the pathogenic TP53 mutations in PCa (70%) are protein-stabilising missense mutations that lead to nuclear accumulation. Other TP53 alterations are truncating mutations (or rarely deep deletions) leading to protein loss [28].
Although multiple studies have shown an association between p53 expression and TP53 mutation, inconsistencies were noted by others. These discrepancies could be attributed to limitations of the IHC assay, including antibodies used for detection or to study cohort selection [8, 9]. But, Gesztes et al [4] confirmed the association between pathogenic TP53 mutations and higher p53 expression, which supports the usage of IHC staining of p53 as a substitute for detecting TP53 mutations.
The focality of TP53 alterations in primary PCa can lead to differences in IHC interpretations or DNA sequencing assays. Since p53 IHC detection depends on the increased half-life of the mutant proteins, proteins with destabilising mutations may escape detection. The lower frequency of TP53 mutations in localised PCa could reduce the likelihood of finding an association with increased p53 expression. Since p53 nuclear accumulation is far more frequent in higher grade carcinomas, performing IHC on all primary PCa at diagnosis is unlikely to establish the expected association [10].
According to the current work, tumour P53 expression was found to be low in 60% of the studied PC cases. These findings are quite similar to those reported by Gesztes et al [4] who found that nearly half (49.8%) of the examined prostatic tumours showed focal p53 expression. But, Shurbaji et al [27] reported that immunoreactivity for p53 was only seen in 21% of their studied cancer cases. This discrepancy was documented in Rejeb et al [22] who showed widely variable rate of p53 expression ranging from 1.1% in localised tumours to 54.7% in surgically treated PCa which may be attributed to differences in the methodology of p53 immunostaining assessment, the differences in the techniques, studied specimens (one section or multiple cores of tumour tissue) and patient’s stage (primary tumours, metastasis).
In the present study, p53 expression was significantly positively correlated with serum PSA level, but no statistically significant relation was found between its expression and any of the studied clinicopathological variables or histopathological findings, which could be related to the small sample size. On the contrary, there was statistically significant relation between P53 expression and MSH2 expression denoting its association with high-grade tumours and poor prognosis.
Gesztes et al [4] also found that high p53 expression was significantly associated with distant metastasis and lymph vascular invasion. Moreover, Shurbaji et al [27] reported that immunoreactivity for p53 was strongly related to progression of PCa in the form of grading and staging denoting that inactivation of p53 is a late event during PCa progression.
Conclusion
MMR protein loss, especially MSH2, was seen in a significant subset of PCa. Interestingly, it was associated with significantly higher levels of serum PSA and p53. Moreover, it may be associated with poor prognostic features as large tumour size, higher grade grouping and finally nodal and distant metastasis.
Conflicts of interest
The authors did not have any conflicts of interest.
Funding
The authors declare that they have no fundings.
Author contributions
First and corresponding author: the idea of the research, protocol writing, data collection and paper writing and revision.
Second author: idea participation, providing clinical data cases and writing their results.
Third author: performing serological tests, writing their results and statistical analyses of their data.
Fourth author: idea participation, data collection, writing the background and discussion.
References
1. Barata P, Agarwal N, and Nussenzveig R, et al (2020) Clinical activity of pembrolizumab in metastatic prostate cancer with microsatellite instability high (MSI-H) detected by circulating tumor DNA J Immunother Cancer 8(2) e001065 https://doi.org/10.1136/jitc-2020-001065 PMID: 32788235 PMCID: 7422632
2. Bhat G, Ahamed MA, and Saroha N, et al (2023) Clinicopathological correlation of p53 expression in benign prostatic hyperplasia and prostate adenocarcinoma Indian J Pathol Oncol 9(1) 60–64
3. Li D, Zhan Y, and Wang N, et al (2023) ETV4 mediates dosage-dependent prostate tumor initiation and cooperates with p53 loss to generate prostate cancer Sci Adv 9 eadc9446 https://doi.org/10.1126/sciadv.adc9446 PMID: 37018402 PMCID: 10075989
4. Gesztes W, Schafer C, and Young D, et al (2022) Focal p53 protein expression and lymphovascular invasion in primary prostate tumors predict metastatic progression Sci Rep 12(1) 5404 https://doi.org/10.1038/s41598-022-08826-5 PMID: 35354846 PMCID: 8967869
5. Gillessen S, Armstrong A, and Attard G, et al (2022) Management of patients with advanced prostate cancer: report from the Advanced Prostate Cancer Consensus Conference 2021 Eur Urol 82 115–1141 https://doi.org/10.1016/j.eururo.2022.04.002 PMID: 35450732
6. Graff JN, Alumkal JJ, and Drake CG, et al (2016) Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer Oncotarget 7 52810–52817 https://doi.org/10.18632/oncotarget.10547 PMID: 27429197 PMCID: 5288150
7. Grindedal EM, Zucknick M, and Stormorken A, et al (2024) Outcomes of 10 years of PSA screeningfor prostate cancer in Norwegian men with Lynch syndrome Prostate 84 945–953 https://doi.org/10.1002/pros.24711 PMID: 38629217
8. Guedes LB, Almutairi F, and Haffner MC, et al (2017) Analytic, preanalytic, and clinical validation of p53 IHC for detection of TP53 missense mutation in prostate cancer Clin Cancer Res 23(16) 4693–4703 https://doi.org/10.1158/1078-0432.CCR-17-0257 PMID: 28446506 PMCID: 5559307
9. Guedes LB, Antonarakis ES, and Schweizer MT, et al (2017) MSH2 loss in primary prostate cancer Clin Cancer Res 23 6863–6874 https://doi.org/10.1158/1078-0432.CCR-17-0955 PMID: 28790115 PMCID: 5690834
10. Haffner MC, Mosbruger T, and Esopi DM, et al (2013) Tracking the clonal origin of lethal prostate cancer J Clin Investig 123 4918–4922 https://doi.org/10.1172/JCI70354 PMID: 24135135 PMCID: 3809798
11. Harmon SA, Gesztes W, and Young D, et al (2021) Prognostic features of biochemical recurrence of prostate cancer following radical prostatectomy based on multiparametric MRI and immunohistochemistry analysis of MRI-guided biopsy specimens Radiology 299(3) 613–623 https://doi.org/10.1148/radiol.2021202425 PMID: 33847515 PMCID: 8165944
12. Javeed S, Chughtai A, and Zafar G, et al (2022) An evaluation of the immunohistochemical expression of mismatch repair proteins (MSH2, MSH6, MLH1, and PMS2) in prostate adenocarcinoma Cureus 14(7) e27448 PMID: 36051725 PMCID: 9420449
13. Kagawa M, Kawakami S, and Yamamoto A, et al (2021) Prevalence and clinicopathological/molecular characteristics of mismatch repair protein-deficient tumours among surgically treated patients with prostate cancer in a Japanese hospital-based population Jpn J Clin Oncol 51(4) 639–645 https://doi.org/10.1093/jjco/hyaa207
14. Ku SY, Rosario S, and Wang Y, et al (2017) Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance Science 355(6320) 78–83 https://doi.org/10.1126/science.aah4199 PMID: 28059767 PMCID: 5367887
15. Le DT, Uram JN, and Wang H, et al (2015) PD-1 blockade in tumors with mismatch-repair deficiency N Engl J Med 372 2509–2520 https://doi.org/10.1056/NEJMoa1500596 PMID: 26028255 PMCID: 4481136
16. Li QK, Lih TM, and Wang Y, et al (2022) Improving the detection of aggressive prostate cancer using immunohistochemical staining of protein marker panels Am J Cancer Res 12(3) 1323–1336 PMID: 35411226 PMCID: 8984898
17. Mucci LA, Hjelmborg JB, and Harris JR, et al (2016) Familial risk and heritability of cancer among twins in Nordic countries JAMA 315 68–76 https://doi.org/10.1001/jama.2015.17703 PMID: 26746459 PMCID: 5498110
18. Oka S, Urakami S, and Hagiwara K, et al (2023) The prevalence of lynch syndrome (DNA mismatch repair protein deficiency) in patients with primary localized prostate cancer using immunohistochemistry screening Hered Cancer Clin Pract 21 20 https://doi.org/10.1186/s13053-023-00265-1 PMID: 37828628 PMCID: 10568829
19. Pejčić T, Todorović Z, and Đurašević S, et al (2023) Mechanisms of prostate cancer cells survival and their therapeutic targeting Int J Mol Sci 24(3) 2939 https://doi.org/10.3390/ijms24032939
20. Qasim A, Zafar A, and Batool S, et al (2021) Screening of endometrioid adenocarcinoma and atypical hyperplasia for mismatch repair genes by immunohistochemistry: a single institute experience Pak J Pathol 32(2) 61–66
21. Quinn DI, Stricker PD, and Kench JG, et al (2019) P53 nuclear accumulation as an early indicator of lethal prostate cancer Br J Cancer 121(7) 578–583 https://doi.org/10.1038/s41416-019-0549-8 PMID: 31409910 PMCID: 6889144
22. Rejeb B, Kouki N, and Elfekih S, et al (2024) Prognostic significance of tumor suppressor protein p53 in prostate cancer La Tunisie Medicale 102(2) 111–115
23. Rodrigues DN, Rescigno P, and Liu D, et al (2018) Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer J Clin Invest 128 4441–4453 https://doi.org/10.1172/JCI121924
24. Schweizer MT, Cheng HH, and Tretiakova MS, et al (2016) Mismatch repair deficiency may be common in ductal adenocarcinoma of the prostate Oncotarget 7 82504–82510 https://doi.org/10.18632/oncotarget.12697 PMID: 27756888 PMCID: 5347709
25. Sekhoacha M, Riet K, and Motloung P, et al (2022) Prostate cancer review: genetics, diagnosis, treatment options, and alternative approaches Molecules 27(17) 5730 https://doi.org/10.3390/molecules27175730 PMID: 36080493 PMCID: 9457814
26. Sharma M, Yang Z, and Miyamoto H (2020) Loss of DNA mismatch repair proteins in prostate cancer Medicine 99(19) e20124 https://doi.org/10.1097/MD.0000000000020124 PMID: 32384491 PMCID: 7220555
27. Shurbaji MS, Kalbfleisch JH, and Thurmond TS (1995) Immunohistochemical detection of p53 protein as a prognostic indicator in prostate cancer Hum Pathol 26(1) 106–109 https://doi.org/10.1016/0046-8177(95)90122-1 PMID: 7821906
28. Stopsack KH, Salles DC, and Vaselkiv JB, et al (2023) P53 immunohistochemistry to identify very high-risk primary prostate cancer: a prospective cohort study with three decades of follow-up Eur Urol Oncol 6(1) 110–112 https://doi.org/10.1016/j.euo.2021.12.003
29. Teroerde M, Nientiedt C, and Duensing A, et al (2021) Revisiting the role of p53 in prostate cancer Prostate cancer eds SRJ Bott and KL Ng (Brisbane: Exon Publications) pp 113–124
30. Van Leenders GJ, Van Der Kwast TH, and Grignon DJ, et al (2020) The 2019 International Society of Urological Pathology (ISUP) consensus conference on grading of prostatic carcinoma Am J Surg Pathol 44(8) e87 https://doi.org/10.1097/PAS.0000000000001497 PMID: 32459716 PMCID: 7382533
31. Wilczak W, Rashed S, and Hube-Magg C, et al (2017) Up-regulation of mismatch repair genes MSH6, PMS2 and MLH1 parallels development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer Carcinogenesis 38 19–27 https://doi.org/10.1093/carcin/bgw116
32. Yan H, Wu Y, and Cui X, et al (2022) From cognitive MR-targeted fusion prostate biopsy to radical prostatectomy: incidence and predictors of gleason grade group upgrading in a Chinese cohort Biomed Res Int 2022 7944342 https://doi.org/10.1155/2022/7944342 PMID: 36033582 PMCID: 9402296
33. Zhang H, Yang X, and Xie J, et al (2023) Clinicopathological and molecular analysis of microsatellite instability in prostate cancer: a multi-institutional study in China Front Oncol 13 1277233 https://doi.org/10.3389/fonc.2023.1277233 PMID: 37901334 PMCID: 10613026
34. Zhou M (2018) High-grade prostatic intraepithelial neoplasia, PIN-like carcinoma, ductal carcinoma, and intraductal carcinoma of the prostate Mod Pathol 31(1) 71–79 https://doi.org/10.1038/modpathol.2017.138
35. Hashemi M, Azizi F, and Najmabady NA, et al (2024) Epidemiology, risk factors and histopathological profile of prostate cancer Prostate Cancer: Molecular Events and Therapeutic Modalities eds G Sethi, M Ashrafizadeh, and N Ebrahimi (Singapore: Springer) https://doi.org/10.1007/978-981-97-4612-5_2