Clinical significance of microRNA-200 and let-7 families expression assessment in patients with ovarian cancer
Severyn Ferneza1, Markiyan Fetsych1, Roman Shuliak2, Halyna Makukh3, Natalia Volodko1, Roman Yarema1 and Taras Fetsych1
1Department of Oncology and Radiology FPGE, Danylo Halytsky Lviv National Medical University, Hasheka 2A str., Lviv 79000, Ukraine
2Department of Microinvasive Surgery, Lviv State Regional Oncology Treatment and Diagnostic Center, Hasheka 2A str., Lviv 79000, Ukraine
3Institute of Hereditary Pathology, National Academy of Medical Sciences of Ukraine, Lviv 79000, Ukraine
Abstract
Ovarian cancer (OC) represents the most lethal malignancy in gynaecologic oncology practice and shows a high recurrence rate due to its early chemoresistance to first-line chemotherapy. Yet, timely selection of the correct treatment strategy is likely to prolong a patient’s survival. MicroRNAs (miRNAs) are a class of short non-coding RNAs responsible for the expression of 30%–60% of human genes. In numerous studies, miRNAs have been used to provide the overall prognosis for patients and analyse the process’s prevalence and responses to chemotherapy. In particular, miRNAs as markers for predicting the sensitivity of OC to platinum- and taxane-based chemotherapeutics can significantly improve the treatment efficacy. This article highlights two families of miRNAs: miR-200 and let-7, which are promising for further research on OC and its chemosensitivity.
Keywords: microRNA, ovarian cancer, biomarkers
Correspondence to: Severyn Ferneza
Email: severyn.ferneza@gmail.com
Published: 14/06/2021
Received: 20/11/2020
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Today, ovarian cancer (OC) ranks among all cancers as the 7th leading cause of morbidity and mortality: 6.6 per 100,000 women are diagnosed with OC; 3.9 per 100,000 women die from OC [1, 2]. OC is the 5th leading cause of death in gynaecologic oncology patients [1].
Early-stage OC is highly curable, which is supported by the high 5-year survival rate of such patients [3]. While the 5-year survival rate makes up 29%, OC is diagnosed in the late stages in most cases. Only 15% of cases are diagnosed in the first stage when the 5-year survival rate reaches 92% [2]. The overall 5-year survival does not exceed 45%–47% [2] and is characterised by the high recurrence rate – up to 20%–25% on the stages I–II, and up to 70% when the disease is discovered in the late stages of its development. About 25% of all relapses occur within 6 months after the treatment completion; such ovarian tumours are considered chemoresistant and have an unfavourable prognosis [3]. Therefore, quick and reliable cancer prognostic approaches are essential at this stage.
The major scientific achievement in the OC study of the past decades was the definition of OC as a heterogeneous group of diseases within one morphological variant. By this definition, all epithelial tumours fall into two groups based on the course and prognosis of the disease, namely: type I – with more favourable prognosis (low-grade serous and endometrioid carcinomas, clear cell and mucinous OC, Brenner’s tumour), and type II with less favourable prognosis (high-grade serous OC, undifferentiated carcinoma and mesodermal tumours) [4]. This discovery was based on molecular genetics and later was implemented in clinical practice. In 2014, WHO and International Federation of Gynecology and Obstetrics (FIGO) revised its classification and updated it by classifying the serous OC as high and low malignant [5]. Hence, each of them was characterised by different genetic mutations, response to chemotherapy and differential prognosis [4]. An advanced analysis of the genome and transcriptome in OC samples, within the Cancer Genome Atlas project, allowed to outline four transcriptional subtypes of high-grade OC (mesenchymal, immunoreactive, differentiated and proliferative), which differ by the duration of patient's survival and the response to treatment three subtypes of microRNAs (miRNAs) [6]. In this respect, the results of the molecular genetic research linking miRNA and OC progression can be crucial for the development, implementation and efficiency assessment of the new treatment methods and the selection of patients for their application [7].
Discovered recently, miRNAs are known as a class of short non-coding RNAs (22 nucleotides). miRNAs can be found in tumour tissue and body fluids and regulate the expression of around 30% of human genes [8]. miRNAs are able to regulate the tumour’s response to treatment and, accordingly, affect the prognosis for a patient. According to recent studies, the expression level of some miRNAs affects the tumour resistance to chemotherapy [8] and radiation therapy [9]. Although a significant volume of data supports the miRNA role as an indicator for cancer prognosis during treatment [8], more information and methodology are needed to describe this phenomenon fully. For example, miRNAs within one family often show different properties in the process of oncogenesis, and their expression in blood and tumour tissue requires more detailed study. miRNAs of two families: miR-200 and let-7 are most frequently discussed in the publications on OC topic. Thus, the purpose of this review is to summarise and analyse the results of these studies and evaluate the possibility of clinical application of the specific miRNA expression.
Role of microRNA-200 and let-7 members in clinical outcome
miRNA correlates with cancer progression
microRNA-200 family
Out of all miRNAs known up to now, miRNA-200 family has been proven to play an essential role in carcinogenesis regulation. However, there are fundamental differences in how each member of this group functions. The family members in the miRNA of group 200 are typically located on two loci of different chromosomes, namely miR-200a, miR-200b, miR-429 are located together on chromosome 1, whereas miR-200c and miR-141 – on chromosome 12 [10]. Hu et al [11] suggest that part of miRNA-200 family, which are located on chromosome 1, has the possibility of co-expression and regulation. It is likely that miRs located on chromosome 1 have a similar impact on OC carcinogenesis. It may also appear that miRNA-200 family members located on chromosome 12 might have a similar tendency to co-regulation and co-expression.
miRNA-200 family is responsible for managing epithelial-mesenchymal transition (EMT) and its reverse process – mesenchymal-epithelial transition (MET), which can lose their normal functioning and lead to carcinogenesis under influence of dysregulated miRs. Zinc finger E-box-binding homeobox 1 (ZEB1) and Zinc finger E-box-binding homeobox 2 (ZEB2) genes are directly responsible for regulation of EMT and MET processes via downregulation of E-cadherin – the facilitator of intercellular binding and cell migration [12]. miR-200 family and ZEB1/ZEB2 control each other via mutual negative feedback – a regulatory mechanism by which miRNA-200 level can downregulate the two genes and vice versa [13]. The disruption of this process can lead to the failure in cell plasticity and develop into cancer metastasis. Interestingly, under some circumstances, miR-200c can downregulate cell proliferation, migration and invasion despite low E-cadherin levels [14]. In experiments on mice, a high level of miR-200c expression at the initial stages of tumour formation led to a significantly better tumour regression than after restoring the miR-200c levels in already formed tumour [15]. The expression of miR-200c was also observed in the inflammatory component of the cell population, which proves its involvement in the regulation of inflammation and, consequently, invasion [12]. Depending on the location of the HuR RNA-binding protein in the miR-200c cell, miR-200c indirectly alters its function by stimulating or inhibiting Tubulin beta chain 3 gene (TUBB3) expression [16]. Therefore, in some patients with a high level of miR-200c and TUBB3, the prognosis was worse: both recurrence-free period and overall survival were shorter. Such mechanism of miRNA interaction with cellular proteins may explain the differences in the function of the same miRNAs concluded by different studies [16].
This statement was partially supported in several studies with tumour tissues and cell lines. However, there is no consensus on the role of miR-200a in the progression of OC. Several studies revealed an increased level of miR-200a tissue expression at the advanced stages (III–IV) of OC [17–19]. Instead, some other studies have shown that miR-200a expression decreases with the disease progression [20]. Suo et al [21] observed a significant increase of miRNA expression in patients with OC but did not provide any data showing the change in expression depending on the stage of the disease. Interestingly, miR-200a expression is present not only in OC-affected tissues. Its level is higher in comparison to tissues taken from healthy individuals [18] and benign ovarian tumours [13]. Similar observations were made about miR-200a expression in lymphatic tissues. Specifically, decreased level of miR-200a was observed in intact lymph nodes and increased – in metastatic lymph nodes [18, 21], indicating the miR-200a potential as a carcinogenic miRNA. In the same time, in several studies, higher miR-200a expression was characteristic of patients without lymph node metastases [13, 20] and increased – in patients with ascites occurrence [20]. These discrepancies are most likely caused by the differences in preservation technique used in each study: frozen tissue samples in some [13] and formalin-fixed paraffin-embedded (FFPE) OC tissue preservation in other studies [20]. One group does not provide information of how tissue samples were stored during the study. There is a high chance of miRNA degradation in formalin samples. Therefore, choosing the appropriate sample preservation method may be crucial for further analysis [22].
Among studied publications, very little data was found on the correlation between the miR-200c tumour tissue levels and the stage of OC progression. For instance, one group has stated that the expression of miR-200c is higher at stages I–II than stages III–IV and has a lower incidence of lymph node involvement [23]. On the contrary, another group has observed an increased level of miR-200c at the stages III–IV [19].
The expression of miR-200b in OC cells is higher as compared to healthy cells [24]. Different results were obtained from patients’ tumour tissue – it was noticed that miR-200b is overexpressed in the tumour tissue of patients with stage I OC and has low expression in stage III [25]. This proves miR-200b to be oncosuppressive miR, and its drop leads to cancer progression. Such differences in statements are most likely caused by different research material used in both studies.
Unlike the healthy cells, those affected by OC showed higher expression of miR-141 and miR-429 [26]. This statement was proved by the experiments on tumour tissue samples [11].
The level of miRNA-200 family may be a prognostic factor for patient survival. In particular, an increased tissue expression of miR-200a correlated with the lower overall survival and relapse-free period [19]. Simultaneous overexpression of miR-200a, c, b was observed in women with lower overall survival [27]. In the same time, hyperexpression of miR-200a appears to be typical in the case of overall and recurrence-free survival [25, 27]. Complementary, the patients with low tissue expression of miR-200a had lower overall survival [11, 20] and shorter recurrence-free survival [11], supporting the above studies (Table 1). Besides these results, other groups have reported reduced [13, 17] or increased [28, 29] tissue expression of miR-200a but did not investigate the correlation between the miRNA levels and patient survival. Finally, some investigations found no relationship between the miR-200a expression level and patient survival [28, 30]. Overall, these findings suggest that miR-200a expression patterns in most cases have been indicative of a carcinogenic process and may prove to have a clinical value for patient’s prognosis (Table 2). Yet, more research on this topic is required.
In many studies, high miR-200c expression in tumour tissue or cell lines has been observed [28, 31]. Other authors, however, indicate a decrease in miRNA expression [32, 33]. Also, the results regarding the dependence of the miR-200c expression level on the prognosis of patients are contradictory: some sources show that miR-200c expression is beneficial for patient survival (Table 3) [28, 31], while the others state the opposite (Table 4) [19, 31] One research group determined miRNA expression of miR-200c-3p only in women with high-grade serous and clear cell histological subtype of OC. They found a negative inverse correlation between miR-200c-3p level and prognosis for patients only in the serous subtype group [34]. These findings show the potential involvement of the miR-200c in cancer progression but require more detailed evidence to follow.
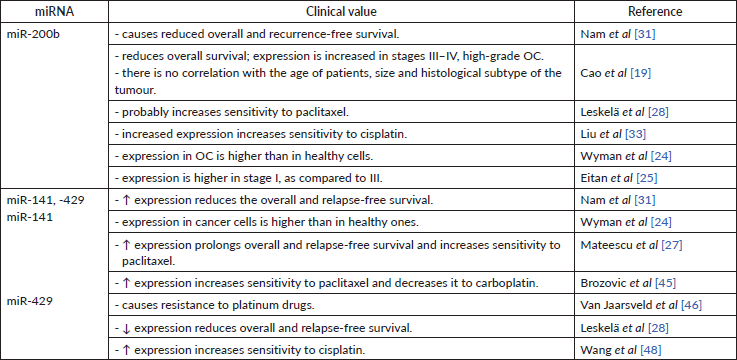
High miR-200b levels in serum [35] and tissue [19, 31] are associated with lower overall and relapse-free survival. These findings strictly indicate miR-200b as carcinogenic miR (Table 5).
Overexpression of miR-141 and miR-429 may also serve as a marker for the prediction of reduced recurrence-free and overall survival [31]. Another study supported this statement, in which higher expression of miR-429 in OC cells compared to healthy cells [24]. At the same time, some research indicates that miR-141 and miR-200a can be a positive prognostic factor for patients (Table 5) [27, 28]. Both groups of authors used different techniques to store collected material – this may again serve as a reason for results discrepancies.
Table 1. Data supporting the oncosuppressive role of miR-200a.
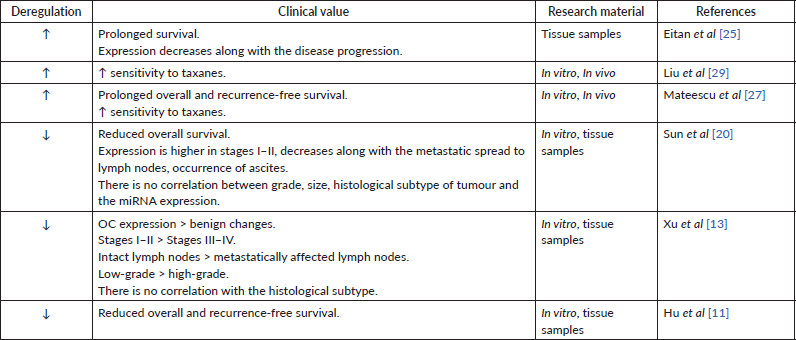
Table 2. Data supporting the carcinogenic role of miR-200a.
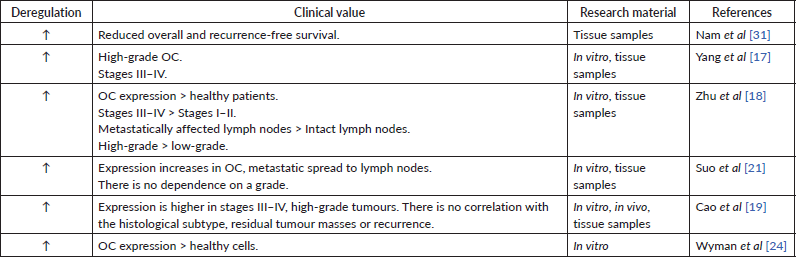
Table 3. Data supporting the oncosuppressive role of miR-200c.
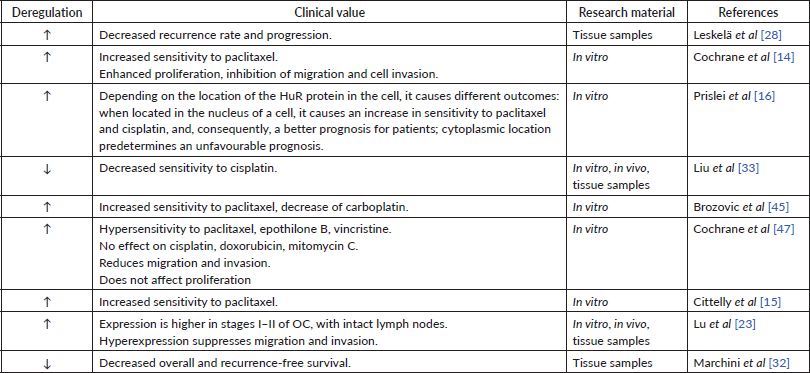
Table 4. Data supporting the carcinogenic role of miR-200c.

No correlation was found between the tumour grade and miR-200 family expression in tumour tissue, except for miR-200b [19]. No relationship was found neither of miR-200a [20, 21] nor of miR-200c, b [19] between miR expression and tumour size. Overexpression of miR-200a was typical for women with high-grade serous OC [17, 18]. At the same time, another group pointed at the reduced expression of miR-200a in patients with high-grade OC as compared to those with low-grade [13]. Such discrepancies may occur due to the different tissue preservation technique. The amount of high- and low-grade groups also remains unclear. The relationship between the histological subtype of OC and miRNA expression also remains controversial. Most studies, however, indicate the absence of such correlation [13, 20]. In contrast, another research [36] have found an increased expression of miR-200a, c in serous, endometrioid and clear cell subtypes. These discrepancies are most likely caused by different tissue preservation technologies: FFPE was used [20], in another – snap frozen tissues [36]. One author does not provide their methodology. miR-200b is hyperexpressed in clear cell OC [34] as well as in its endometrioid and serous subtype [36]. Increased expression of miR-141 is typical for endometrioid and serous subtype of OC [16].
Overall, miR-200 family has diverse roles in carcinogenesis. Sample preservation techniques are the potential primary cause of data discrepancies in miR studies and should be consistent for reliable comparisons.
Let-7 family
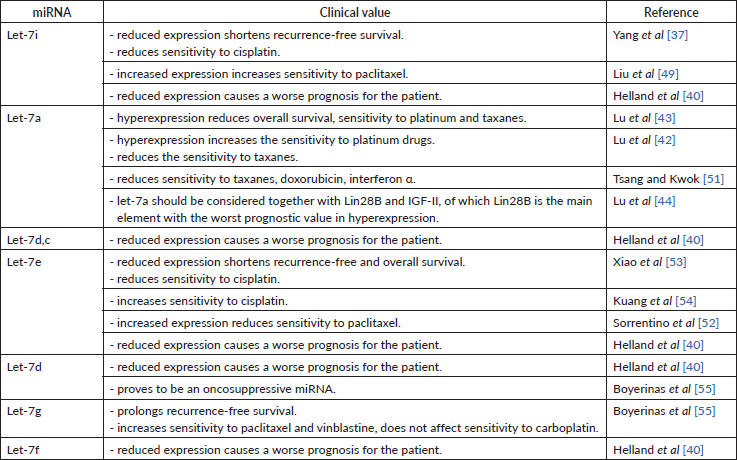
Let-7 family was one of the first miRs discovered to have a suppressive impact on carcinogenesis [37]. The let-7 miRNA family consists of 12 members located at eight loci of seven chromosomes: chromosome 3 – let-7g; 9 – a-1, d, f-1; 11 – a-2; 12 – i; 19 – e; 21 – c; 22 – a-3, b; x – f-2, miR-98 [38]. As compared to the miRNA 200 family, the let-7 family is less studied, but its role in carcinogenesis remains undoubted. Dysregulation of the let-7 family has been found in medulloblastoma, breast cancer, OC, melanoma and non-small cell lung cancer, clearly showing the role of the miRNA family in the carcinogenesis of many malignancies [38]. Decreased expression of let-7e, let-7f, let-7d, let-7c, let-7a-e, let-7i, let-7a, let-7b in tumour samples and decreased let-7f, let-7d, let-7a-e in cell lines [39], decreased cell growth and reduced lymph node involvement [38] proves an oncosuppressive role of this miRNAs family (Table 6).
All studies we have looked into agree on a cancer-suppressive role of let-7i. As it turns out, its low expression in tissue is characteristic of patients with poor prognosis [40], short recurrence-free period [37]. Low let-7i expression in patients with OC [39] might indirectly indicate its oncosuppressive role as well. Despite this, the decreased let-7i expression has been associated with the depth of metastasis invasion into lymph nodes in patients with gastric cancer. It is possible that let-7i may play a similar role in patients with OC [41]. No data could be found to describe the correlation between let-7i expression and disease progression as well as the degree of tumour differentiation in patients with OC. However, being able to negatively regulate the expression of Toll-like receptor 4 (TLR4) and MyD88 genes, let-7i prolongs the periods of overall and progression-free survival [41].
Most researchers observe an increased let-7a expression in ovarian tumour tissue [42, 43]. In contrast, another research group detected low let-7a expression both in vitro and in tumour masses [39]. Both research groups used significantly varying sample sizes in their studies. Furthermore, the first research group did not use cell lines in their research. A number of studies showed that let-7a expression in clinic should be assessed in combination with Lin28B and Insulin-like growth factor 2 (IGF-II), as their simultaneous high expression resulted in more frequent recurrences and higher overall mortality in patients receiving platinum and paclitaxel treatment [44]. This, in turn, may testify about the negative regulation of OC cells sensitivity to drugs of the first-line chemotherapy. High let-7a-3 activity indirectly reduced overall survival by 40%, but this did not affect the progression of OC [44]. High Lin28B expression is a more accurate indicator of poor prognosis than the dysregulation of any other axis component. Low Lin28B expression correlated with the less aggressive OC, lower tumour malignancy and better chemotherapy response, whereas high IGF-II expression correlated with very low survival and sensitivity to paclitaxel, yet let-7a itself did not affect survival [43].
miRNAs modulate chemotherapy response via gene regulation
miRNA-200 family
MiRs may affect tumour drug sensitivity through gene regulation. In particular, it has been determined that overexpression of miR-200a promotes sensitivity to paclitaxel by inhibiting the Mitogen-activated protein kinase 14 (MAPK14) gene and p38α protein [27]. An increased expression of miR-200a in cell lines was associated with taxane sensitivity. Interestingly, that growth in paclitaxel concentration has not significantly changed cellular chemosensitivity [29].
Whereas miR-200a possesses strictly chemosensitising abilities (Table 1), for miR-200c some discrepancies were found. Nevertheless, the level of miR-200c expression undoubtedly impacts the tumour response to platinum and taxanes. In vitro experiments showed that the expression of miR-200c drops 4–5-fold from its normal levels in tissues with observed resistance to paclitaxel. At the same time, miR-200c reduces the sensitivity of cells to carboplatin by increasing the sensitivity to taxanes [45]. This mechanism remains unclear. These results are consistent with those of Van Jaarsveld et al [46], who claim that hyperexpression of miR-200c causes little resistance to cisplatin. In contrast, Liu et al [33] indicate that decreased miR-200c expression correlates with decreased sensitivity to cisplatin in vitro, in vivo, and retrospectively, based on tissue samples from patients with OC. miR-200c inhibits TUBB3, thereby increasing the sensitivity of cells to cisplatin [16] and paclitaxel [14, 16]. This statement was supported in the study using tumour tissue samples – lower miR-200c expression was indicated in patients with a poorer response to first-line OC chemotherapy such as taxanes and platinum-based chemotherapy [28]. Similar mechanism of chemosensitivity regulation was found by another study, where miR-200c increased the efficacy of in vitro drugs, in particular, to paclitaxel (82%–85%), vincristine (33%–35%) and epothilone B (43%–50%), which all affect the microtubule system [47]. Thus, miR-200c evidently has a positive impact on tumour chemosensitivity to taxanes (Table 3). The estimated negative effect to platinum-based drugs could have been caused by the differences in cell lines used in the study by Brozovic et al [45]. It also appears that such discrepancy of miR-200c function might be caused by different position of HuR in cell, also mentioned by Prislei et al [16].
No direct impact on tumour sensitivity to taxanes by miR-200b was found. However, miR-200b has the same impact on class III β-tubulin as any other member of miR-200 family. Thus, it may have an indirect influence on OC sensitivity to taxanes [28]. Another study has determined that increased expression of miR-200b results in better sensitivity to cisplatin (Table 5) [33].
It has been discovered that hyperexpression of miR-141 promotes sensitivity to paclitaxel by inhibiting the MAPK14 gene and the p38α protein [27]. miR-141’s positive effect on OC sensitivity to taxanes was also supported by in vitro research [45]. Alike the miR-200c, miR-141 also reduces cellular sensitivity to carboplatin by increasing sensitivity to taxanes [45]. Decreased expression of miR-141 in vitro led to increased resistance to paclitaxel by 4–5 times, and vice versa – after the administration of miR-141, the sensitivity to paclitaxel [45] has increased. miR-141’s negative effect on cell-to-platinum sensitivity was supported by another group, pointing out that overexpression of miR-141 caused the resistance to platinum-based drugs by inhibiting the expression of Kelch-like ECH-associated protein 1. It appears that a negative feedback loop between cisplatin and miR-141 exists. Cisplatin caused a transient increase in miR-141 expression, the level of which returned to normal in 24 hours [46]. Same as for miR-200c, miR-141 undoubtedly has a positive impact on a chemosensitivity to taxanes (Table 5). The estimated negative effect to platinum-based drugs might be caused by different OC cell lines used in the study.
miR-429 indicates a favourable prognosis for patient treatment – it has been revealed that hyperexpression of miR-429 in vitro leads to increased cell sensitivity to cisplatin (Table 5) [48].
Table 5. The clinical value of mir-200b, -141, -429.
Let-7 family
Some sources define let-7i as a regulator of OC sensitivity to chemotherapeutics. In particular, low levels of let-7i expression in tissue and in vitro cause low sensitivity to cisplatin [37]. It was also observed that progesterone increases the expression of let-7i in SKOV-3 cell lines (ovarian cancer cell lines) and therefore is involved in cell proliferation and chemosensitivity, as concluded by the authors. Introducing let-7i in OVCAR-3 cell lines reduced cell viability by 36% and increased apoptosis by 34% [49]. let-7i included in a chimeric construct with the Mucin 1 aptamer increased the permeability of this miRNA into the cell and led to an increase in the efficiency (sensitivity) of paclitaxel [49]. Radiation therapy reduces the therapeutic effect of let-7 family miRNA observed during chemical treatment [50]. This relation has been discovered for lung cancer and may be consistent in OC as well.
The role of let-7a in OC chemosensitivity was determined in a study conducted by Lu et al [42]. The authors point out that hyperexpression of this miRNA can potentiate the effect of platinum drugs in a mono-regime, but can negatively affect the prognosis upon combination treatment (e.g. first-line carboplatin + paclitaxel). Some studies conclude that let-7a reduces the tumour sensitivity to paclitaxel. This was supported by another research, where low Lin28B expression (from let-7a – Lin28B – IGF-II axis) correlated with the less aggressive OC, lower tumour malignancy and better chemotherapy response. In contrast, high IGF-II expression correlated with low survival and sensitivity to paclitaxel (yet let-7a alone did not affect survival) [43]. Another group observed that let-7a decreases cellular sensitivity to paclitaxel, doxorubicin and interferon alpha by inhibiting caspase-3 and, consequently, decreased cell apoptosis [51]. This study was carried out on breast cancer cells and hepatocellular carcinoma cells. These findings, however, may be relevant in OC as well.
Among the let-7 family members, five are associated with cell culture chemoresistance. All of them are expressed differently depending on cell lines, in particular: let-7e – were increased in paclitaxel-resistant lines but had reduced expression in others. This may signify a negative regulation of let-7e cell sensitivity to taxanes [52]. The role of let-7e in chemosensitivity to cisplatin is confirmed by Xiao et al [53]. The researchers showed reduced let-7e expression in platinum-resistant cell lines and overexpression in platinum-sensitive cells [53]. Let-7e increases the concentration of cisplatin in the cell by 20.2% and inhibits tumour growth in vivo [54]. Let-7e increases cellular sensitivity to cisplatin by inhibiting the Enhancer of zeste homolog 2 gene responsible for Dicer expression, which, in turn, increases sensitivity to cisplatin [54]. The let-7 family also affects the sensitivity to taxanes upon the presence of Mitochondrial inner membrane protease subunit 1 gene – a direct target for let-7 in the cells. High levels of let-7g increase the sensitivity to taxanes and vinblastine, but do not affect the sensitivity to carboplatin. let-7g effects chemosensitivity through the indirect inhibition of Multidrug resistance mutation 1 (MDR1). Patients with high MDR1 expression had a shorter recurrence-free period as compared to those whose MDR1 expression was unchanged or reduced [55]. When MDR is absent, the sensitivity to chemotherapy seems to be independent of let-7g, proving their cumulative effect. Tumour expression of let-7d was lower during re-operation [55], apparently, due to a decrease in tumour mass. Decreased expression of let-7 family members was downregulated in OC, medulloblastoma and breast cancer [38].
Let-7 family members’ expression level is a prominent example of how the miRNAs tissue expression could be used as a predictor of chemotherapeutic response in OC (Table 6). A body of evidence suggests that this effect is mediated by this miRNA close regulation of gene expression. More data is required to fully describe the known and establish other connections between let-7 expression and treatment success.
Extracellular role of miRNA in the assessment of cancer progression and chemosensitivity
miR-200 family
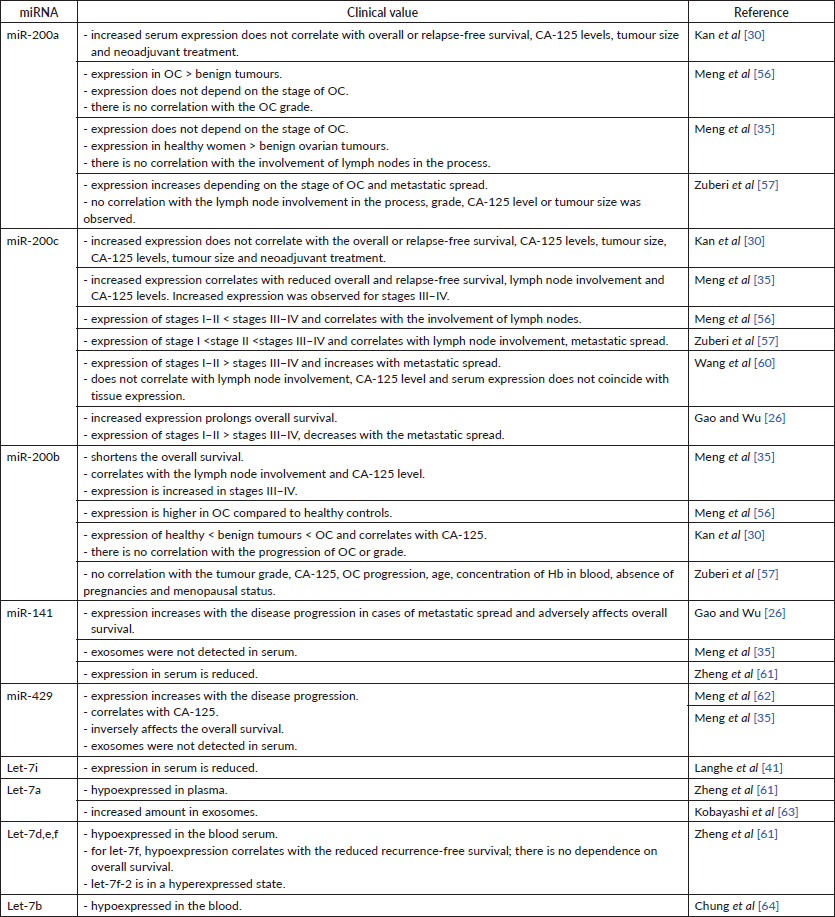
The expression level of miR-200a in serum has been a focus of many studies. All authors reported an increase of miRNA expression in the serum of OC patients independent of the stage [35, 56]. No correlation was established between the miR-200a expression in serum and the duration of overall or recurrence-free survival [30], tumour size [30, 57], serum Cancer antigen 125 (CA-125) [30, 57], the use of neoadjuvant treatment [30], tumour grade [56, 57] or the involvement of lymph nodes in the cancer process [35, 57]. However, there was a correlation between an increased miR-200a expression and the presence of distant metastases and the mucinous subtype of OC [57]. Interestingly, the expression of miR-200a in serum was higher in healthy women in comparision to the group of women with benign ovarian tumours [35].
Table 6. Clinical value of miRNAs of the let-7 family.
Expression of miR-200c is also observed in the serum of patients with OC compared to healthy individuals [30]. Moreover, healthy women can be distinguished from those with OC by the level of serum miR-200c (accuracy – 0.79, and for CA-125 – 50%) [26, 56]. The level of this miRNA itself serves as a factor of the overall survival prediction: high miR-200c levels are observed in patients with longer overall survival [26]. Another research group provides opposite results – high expression level of miR-200c in serum is related to the lower overall survival and to a shorter relapse-free period [35]. No correlation between miRNA levels and patient age, tumour size, neoadjuvant treatment effect, overall survival or recurrence-free survival was found at all [30]. The former group used the whole serum samples in their study, whereas the latter extracted miRs’ exosomes, this could explain the differences in their results. miRNAs are represented in peripheral blood binded with proteins or in exosomes [58]. Thus, measurement of only exosome fraction may not represent the whole extracellular miRNA scope. Ethnical groups also differ by miRNA expression [59]. The number of samples in the one of studies was significantly smaller, which can also affect the overall conclusion.
The level of serum miR-200c correlates with the disease progression. Its expression decreases from stage I to stage IV of OC [26, 47]. Instead, Meng et al [56] and Zuberi et al [57] pointed at a gradual increase in miRNA expression along with the OC stage progression. Another study found no statistically significant increase in miR-200c expression in stages I–II but instead reported advanced stage miRNA overexpression [35]. Serum expression of miR-200c was higher in the case of the formation of secondary foci in the lymph nodes [35, 56], although Wang et al [60] did not observe such a relationship. The level of serum miR-200c was also going up as the remote formation of secondary foci progressed [57, 60]. The results provided by Gao et al [26] oppositely show that the level of serum miRNA was lower in the metastatic than in local OC. Among the above studies, only one reports the correlation between the miR-200c serum level and the CA-125 level [35]. Other researchers did not find such correlation for miRNA expression neither in the serum [30, 60] nor in the tissue [34]. The expression of miR-200c in serum varies depending on the histological subtype of epithelial OC (serous, mucinous, endometrioid, clear cell and undifferentiated) [26]. All these findings show potential role of miR-200c as a target of ‘liquid biopsy’. The reason for discrepancies found in studies most likely hides in different material approach – For instance (f.i.), Wang et al isolated exosomes from the serum, whereas Meng et al and Zuberi et al [57] used the whole serum. As well as nonhomogenous age groups used in studies might have had an impact on miRNA expression.
The expression of miR-200b is also observed in blood serum. Serum expression of miR-200b is sensitive to the ovarian tissue changes: its level is higher in women with benign ovarian tumours compared to healthy ovarian tissue and lower compared to tumour tissues in patients with OC [56]. Opposite results were presented by another study – miR-200b expression was higher in patients with OC [30]. The abundance of miR-200b expression is independent of the stage of the OC progression [56]. Another study shows the potential carcinogenic role of miR-200b by describing its upregulation at stages III–IV of OC and in women with affected lymph nodes [35]. These results were confirmed by Zuberi et al [57]. This study, however, failed to detect the significant increase of miR-200b expression in the serum of OC patients as a function of the disease progression; also, no correlation was found between the miR-200b expression and the level of CA-125, age, tumour grade, tumour size, menopausal status, haemoglobin concentration or absence of pregnancy [57]. Another study denies the correlation between the serum miR-200b levels and tumour grade [56], whereas its correlation with CA-125 levels is established [35]. Again, discrepancies in these reports were most likely caused by the measurements of miR-200b taken in exosomes versus the whole serum.
Both miR-141 and -429 in serum act as carcinogenic miRNAs. Zheng et al [61] have noticed that miR-141 is hypoexpressed in plasma. Another study reported that the serum miR-141 level: i) increases as a function of cancer progression; ii) is higher during metastatic spread and iii) correlates with the lower overall survival [26]. Unlike healthy women’s serum miR-429 levels, those of OC patients increase simultaneously with the disease progression and stage of disease. The serum level of this miRNA is statistically significantly correlated with the level of CA-125 in blood serum. Low expression of miR-429 has been shown to have a positive effect on overall survival, moreover increasing cell migration and invasion [62]. In contrast to this, Meng et al [35] did not detect miR-429 or -141 exosomes in blood serum. The authors measured miRNAs in exosomes, whereas other research groups used the whole serum for it.
Therefore, miR-200 family members, extracellular abundance may be indicative of an optimistic prognosis for the patient (Table 7). Most evidence has been reconciled from blood serum samples, which proves this object reliable for this analysis. Nevertheless, more studies concerning miRNA level in whole serum and exosomes should be done.
Let-7 family
Data on let-7 family expression in serum of the patients with OC is lower compared to the miR-200 family. However, all available results list let-7 blood expression as a positive prognostic factor for patients with OC. Let-7i expression was detected in serum, but the data on this topic is insufficient. In their study, Langhe et al [41] have found that let-7i in the blood is hypoexpressed in patients with OC.
The expression of let-7a in blood plasma was also detected but not fully described. Zheng et al [61] indicated a decreased miRNA expression in plasma [61]. Instead, Kobayashi et al [63] found out a large number of exosomes with high content of let-7 miRNAs, in particular, let-7a, b, c, d, e, f in the in vitro study.
No correlation has been found between let-7f plasma expression and overall survival, however low miRNA level correlated with reduced recurrence-free survival [61]. Decreased expression of let-7b, d, i, c, e, f correlates with the poor prognosis [40]. Low let-7e expression, high Rad51 expression and BRAC1 are associated with unlikely recurrence and overall survival [53].
Zheng et al [61] compared the miRNA expression in plasma and tumour tissue of women with OC and healthy ovarian tissue controls. There was a difference with control in 30 plasma miRNAs, including hypoexpressed: let-7a, d, e, f, miR-98; and hyperexpressed: let-7f-2. Such data is confirmed in another study, where serum let-7b was detected in a hypoexpressed state in patients with OC [64].
Available data states that let-7 family has predominantly oncosuppressive properties – let-7 expression in peripheral blood was indicated as a positive prognostic factor for patients with OC (Table 7).
Table 7. Expression of miRNAs in the blood and their clinical value.
Conclusions
Despite the controversial results of experiments on the role of miRNA families 200 and let-7 in OC, their potential benefit as markers of early OC diagnosis and its prevalence remains encouraging. The miRNA-200 family is presented as a regulator of EMT and its counterpart – MET through impacting ZEB1/ZEB2 expression. Disruption in this process can lead to failure in cell plasticity and promote cancer metastasis. Thus, early detection of miRNA-200 family dysregulation in blood through ‘liquid biopsy’ could be beneficial for the timely OC diagnostic. miR-200 family regulation impact on EMT-MET mechanism in the scope of OC treatment remains to be addressed by further research.
miRNA family of let-7 in the scope of OC is studied insufficiently. Available data states that it has predominantly oncosuppressive properties. Decreased expression of let-7e, f, d, c, a-e, i, a, b in tumour samples and decreased let-7f, d, a-e in cell lines, decreased cell growth and reduced lymph node involvement prove an oncosuppressive role of this miRNAs family. All available data support the oncosuppressive role of the family – let-7 expression in peripheral blood was indicated as positive prognostic factor for patients with OC.
For both miR-200 and let-7 family, data concerning their role in carcinogenesis of OC appeared to be different during the evaluation of blood and tumour tissue expression. F.i. miR-200a and let-7a expression in tumour tissue represent them mostly as carcinogenic miRNAs. However, their expression in serum samples proved their oncosuppressive abilities. miR-200 and let-7 family role discrepancies are caused by various differences in research planning. Research groups used different methods to store biomaterial. This is crucial. F.i. fixation in formalin may hinder miRNA level in tumour tissue. Similarly, measurement of only exosome fraction may not represent the whole extracellular miRNA scope. Furthermore, no absolute ranges for miR-200 and let-7 family expression have been established yet, which hinder the standard evaluation of miRNA expression and its implementation into clinical practice.
The miRNAs of the 200 and let-7 families are promising for their further use in clinical practice, but the mechanisms of their involvement in cancer therapeutic response require additional research. The procedure of miRNA extraction and evaluation should be standardised in order to provide reliable data for clinical use. Future studies in the field, once incorporated these suggestions, may eliminate or explain the existing contradictions concerning the role of these miRNAs in the process of carcinogenesis, prognosis and impact on chemosensitivity in patients with OC. This will complement the current knowledge of OC molecular and cell biology, have an immense impact on the development of accurate, time- and cost-efficient therapeutic progression assessment and motivate new strategies for OC clinical treatment in the future.
List of abbreviations
OC, Ovarian cancer; miRNA, miR, microRNA; FFPE, Formalin-fixed paraffin-embedded; MAPK14, Mitogen-activated protein kinase 14; CA-125, Cancer antigen 125; TUBB3, Tubulin beta chain 3 gene; Lin28B, Lin-28 homolog B; IGF-II, Insulin-like growth factor 2; MDR1, Multidrug resistance mutation 1; EMT, Epithelial-mesenchymal transition; MET, Mesenchymal-epithelial transition; ZEB1, Zinc finger E-box-binding homeobox 1; ZEB2, Zinc finger E-box-binding homeobox 2; f.i., For instance.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Funding statement
The authors received no financial support for the research or authorship of this paper. This publication was supported by ecancer Global Foundation.
References
1. Coburn SB, Bray F, and Sherman ME, et al (2017) International patterns and trends in ovarian cancer incidence, overall and by histologic subtype Int J Cancer 140(11) 2451–2460 https://doi.org/10.1002/ijc.30676 PMID: 28257597 PMCID: 5595147
2. Reid BM, Permuth JB and Sellers TA (2017) Epidemiology of ovarian cancer: a review Cancer Biol Med 14 9–32 https://doi.org/10.20892/j.issn.2095-3941.2016.0084 PMID: 28443200 PMCID: 5365187
3. Ushijima K (2010) Treatment for recurrent ovarian cancer – at first relapse J Oncol 2010 497429 https://doi.org/10.1155/2010/497429
4. Kurman RJ and Shih IM (2010) The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory Am J Surg Pathol 34(3) 433–443 https://doi.org/10.1097/PAS.0b013e3181cf3d79 PMID: 20154587 PMCID: 2841791
5. Prat J (2015) Staging classification for cancer of the ovary, fallopian tube, and peritoneum Obstet Gynecol 126(1) 171–174 [http://journals.lww.com/00006250-201507000-00026] Date accessed: 21/03/21 https://doi.org/10.1097/AOG.0000000000000917 PMID: 26241270
6. Bell D, Berchuck A, and Birrer M, et al (2011) Integrated genomic analyses of ovarian carcinoma Nature 474(7353) 609–615 [http://www.drugbank.ca] Date accessed: 21/03/21 https://doi.org/10.1038/nature10166
7. Hollis RL and Gourley C (2016) Genetic and molecular changes in ovarian cancer Cancer Biol Med 13(2) 236–247 [http://pmc/articles/PMC4944549/] Date accessed: 21/03/21 https://doi.org/10.20892/j.issn.2095-3941.2016.0024 PMID: 27458531 PMCID: 4944549
8. Frederick P, Green H, and Huang J, et al (2013) Chemoresistance in ovarian cancer linked to expression of microRNAs Biotech Histochem 88(7) 403–409 https://doi.org/10.3109/10520295.2013.788736 PMID: 23672416
9. Ke G, Liang L, and Yang JM, et al (2013) MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene Oncogene 32(25) 3019–3027 [www.nature.com/onc] Date accessed: 21/03/21 https://doi.org/10.1038/onc.2012.323
10. Larrea E, Sole C, and Manterola L, et al (2016) New concepts in cancer biomarkers: circulating miRNAs in liquid biopsies Int J Mol Sci 17(5) 627. https://doi.org/10.3390/ijms17050627 PMCID: 4881453
11. Hu X, Macdonald DM, and Huettner PC, et al (2009) A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer Gynecol Oncol 114(3) 457–464 [http://www.ncbi.nlm.nih.gov/pubmed/19501389] Date accessed: 11/03/20 https://doi.org/10.1016/j.ygyno.2009.05.022 PMID: 19501389
12. Ibrahim FF, Jamal R, and Syafruddin SE, et al (2015) MicroRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer J Ovarian Res 8(1) 56 [http://ovarianresearch.biomedcentral.com/articles/10.1186/s13048-015-0186-7] Date accessed: 15/04/20 https://doi.org/10.1186/s13048-015-0186-7 PMID: 26260454 PMCID: 4531514
13. Xu S, Xu P, and Wu W, et al (2014) The biphasic expression pattern of miR-200a and E-cadherin in epithelial ovarian cancer and its correlation with clinicopathological features Curr Pharm Des 20(11) 1888–1895 https://doi.org/10.2174/13816128113199990523
14. Cochrane DR, Howe EN, and Spoelstra NS, et al (2010) Loss of miR-200c: a marker of aggressiveness and chemoresistance in female reproductive cancers J Oncol 2010 821717 [http://www.ncbi.nlm.nih.gov/pubmed/20049172] Date accessed: 20/03/20 https://doi.org/10.1155/2010/821717 PMID: 20049172 PMCID: 2798671
15. Cittelly DM, Dimitrova I, and Howe EN, et al (2012) Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel Mol Cancer Ther 11(12) 2556–2565 https://doi.org/10.1158/1535-7163.MCT-12-0463 PMID: 23074172 PMCID: 3519949
16. Prislei S, Martinelli E, and Mariani M, et al (2013) MiR-200c and HuR in ovarian cancer BMC Cancer 13 72 [https://pubmed.ncbi.nlm.nih.gov/23394580/] Date accessed: 08/04/21 https://doi.org/10.1186/1471-2407-13-72 PMID: 23394580 PMCID: 3576328
17. Yang H, Kong W, and He L, et al (2008) MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN Can Res 68(2) 425–433 [http://www.ncbi.nlm.nih.gov/pubmed/18199536] Date accessed: 11/02/20 https://doi.org/10.1158/0008-5472.CAN-07-2488
18. Zhu CL and Gao GS (2014) miR-200a overexpression in advanced ovarian carcinomas as a prognostic indicator Asian Pac J Cancer Prev 15(20) 8595–8601 [https://www.readcube.com/articles/10.7314%2Fapjcp.2014.15.20.8595] Date accessed: 08/04/21 https://doi.org/10.7314/APJCP.2014.15.20.8595 PMID: 25374174
19. Cao Q, Lu K, and Dai S, et al (2014) Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer Int J Clin Exp Pathol 7(5) 2392–2401 PMID: 24966949 PMCID: 4069884
20. Sun Q, Zou X, and Zhang T, et al (2014) The role of miR-200a in vasculogenic mimicry and its clinical significance in ovarian cancer Gynecol Oncol 132(3) 730–738 https://doi.org/10.1016/j.ygyno.2014.01.047 PMID: 24503464
21. Suo HB, Zhang KC and Zhao J (2018) MiR-200a promotes cell invasion and migration of ovarian carcinoma by targeting PTEN Eur Rev Med Pharmacol Sci 22(13) 4080–4089 [https://pubmed.ncbi.nlm.nih.gov/30024595/] Date accessed: 08/04/21 PMID: 30024595
22. Boisen MK, Dehlendorff C, and Linnemann D, et al (2015) MicroRNA expression in formalin-fixed paraffin-embedded cancer tissue: identifying reference MicroRNAs and variability BMC Cancer 15(1) 1024 [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-015-2030-2] Date accessed: 29/03/21 https://doi.org/10.1186/s12885-015-2030-2 PMID: 26714641 PMCID: 4696166
23. Lu YM, Shang C, and Ou YL, et al (2014) miR-200c modulates ovarian cancer cell metastasis potential by targeting zinc finger E-box-binding homeobox 2 (ZEB2) expression Med Oncol 31(8) 1–11 https://doi.org/10.1007/s12032-014-0134-1
24. Wyman SK, Parkin RK, and Mitchell PS, et al (2009) Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries PLoS One 4(4) e5311 [https://dx.plos.org/10.1371/journal.pone.0005311] Date accessed: 05/05/20 https://doi.org/10.1371/journal.pone.0005311 PMID: 19390579 PMCID: 2668797
25. Eitan R, Kushnir M, and Lithwick-Yanai G, et al (2009) Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients Gynecol Oncol 114(2) 253–259 https://doi.org/10.1016/j.ygyno.2009.04.024 PMID: 19446316
26. Gao YC and Wu J (2015) MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer Tumor Biol 36(6) 4843–4850 [http://www.ncbi.nlm.nih.gov/pubmed/25636451] Date accessed: 21/03/20 https://doi.org/10.1007/s13277-015-3138-3
27. Mateescu B, Batista L, and Cardon M, et al (2011) MiR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response Nat Med 17(12) 1627–1635 [http://www.ncbi.nlm.nih.gov/pubmed/22101765] Date accessed: 23/02/20 https://doi.org/10.1038/nm.2512 PMID: 22101765
28. Leskelä S, Leandro-García LJ, and Mendiola M, et al (2011) The miR-200 family controls β-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients Endocr Relat Cancer 18(1) 85–95 https://doi.org/10.1677/ERC-10-0148
29. Liu N, Zhong L, and Zeng J, et al (2015) Upregulation of microRNA-200a associates with tumor proliferation, CSCs phenotype and chemosensitivity in ovarian cancer Neoplasma 62(4) 550–558 [https://pubmed.ncbi.nlm.nih.gov/25997962/] Date accessed: 08/04/21 https://doi.org/10.4149/neo_2015_066 PMID: 25997962
30. Kan CWS, Hahn MA, and Gard GB, et al (2012) Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer BMC Cancer 12 627 https://doi.org/10.1186/1471-2407-12-627
31. Nam EJ, Yoon H, and Kim SW, et al (2008) MicroRNA expression profiles in serous ovarian carcinoma Clin Cancer Res 14(9) 2690–2695 https://doi.org/10.1158/1078-0432.CCR-07-1731 PMID: 18451233
32. Marchini S, Cavalieri D, and Fruscio R, et al (2011) Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections Lancet Oncol 12(3) 273–285 https://doi.org/10.1016/S1470-2045(11)70012-2 PMID: 21345725
33. Liu J, Zhang X, and Huang Y, et al (2019) mir-200b and mir-200c co-contribute to the cisplatin sensitivity of ovarian cancer cells by targeting DNA methyltransferases Oncol Lett 17(2) 1453–1460 PMID: 30675199 PMCID: 6341622
34. Vilming Elgaaen B, Olstad OK, and Haug KBF, et al (2014) Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker BMC Cancer 14(1) 80 [http://bmccancer.biomedcentral.com/articles/10.1186/1471-2407-14-80] Date accessed: 16/04/20 https://doi.org/10.1186/1471-2407-14-80 PMID: 24512620 PMCID: 3928323
35. Meng X, Müller V, and Milde-Langosch K, et al (2016) Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer Oncotarget 7(13) 16923–35 https://doi.org/10.18632/oncotarget.7850 PMID: 26943577 PMCID: 4941360
36. Iorio MV, Visone R, and di Leva G, et al (2007) MicroRNA signatures in human ovarian cancer Cancer Res 67(18) 8699–8707 https://doi.org/10.1158/0008-5472.CAN-07-1936 PMID: 17875710
37. Yang N, Kaur S, and Volinia S, et al (2008) MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer Cancer Res 68(24) 10307–10314 [http://www.ncbi.nlm.nih.gov/pubmed/19074899] Date accessed: 13/10/19 https://doi.org/10.1158/0008-5472.CAN-08-1954 PMID: 19074899 PMCID: 2762326
38. Wang Y, Hu X, and Greshock J, et al (2012) Genomic DNA copy-number alterations of the let-7 family in human cancers PLoS One 7(9) e44399 https://doi.org/10.1371/journal.pone.0044399 PMID: 22970210 PMCID: 3435307
39. Dahiya N, Sherman-Baust CA, and Wang TL, et al (2008) MicroRNA expression and identification of putative miRNA targets in ovarian cancer PLoS One 3(6) e2436 [https://dx.plos.org/10.1371/journal.pone.0002436] Date accessed: 05/05/20 https://doi.org/10.1371/journal.pone.0002436 PMID: 18560586 PMCID: 2410296
40. Helland Å, Anglesio MS, and George J, et al (2011) Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian cancers PLoS One 6(4) e18064 https://doi.org/10.1371/journal.pone.0018064 PMID: 21533284 PMCID: 3076323
41. Langhe R, Norris L, and Saadeh FA, et al (2020) A novel serum microRNA panel to discriminate benign from malignant ovarian disease Cancer Lett 356(2) 628–636 [http://www.ncbi.nlm.nih.gov/pubmed/25451316] Date accessed: 03/03/20 https://doi.org/10.1016/j.canlet.2014.10.010
42. Lu L, Schwartz P, and Scarampi L, et al (2011) MicroRNA let-7a: a potential marker for selection of paclitaxel in ovarian cancer management Gynecol Oncol 122(2) 366–371 [https://pubmed.ncbi.nlm.nih.gov/21571355/] Date accessed: 08/04/21 https://doi.org/10.1016/j.ygyno.2011.04.033 PMID: 21571355
43. Lu L, Katsaros D, and Canuto EM, et al (2016) LIN-28B/let-7a/IGF-II axis molecular subtypes are associated with epithelial ovarian cancer prognosis Gynecol Oncol 141(1) 121–127 https://doi.org/10.1016/j.ygyno.2015.12.035 PMID: 26751131
44. Lu L, Katsaros D, and Rigault De La Longrais IA, et al (2007) Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis Cancer Res 67(21) 10117–10122 [http://www.ncbi.nlm.nih.gov/pubmed/17974952] Date accessed: 06/05/20 https://doi.org/10.1158/0008-5472.CAN-07-2544 PMID: 17974952
45. Brozovic A, Duran GE, and Wang YC, et al (2015) The miR-200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR-3 and MES-OV cells Mol Oncol 9(8) 1678–1693 https://doi.org/10.1016/j.molonc.2015.04.015 PMID: 26025631 PMCID: 4788969
46. Van Jaarsveld MT, Helleman J, and Boersma AW, et al (2013) miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells Oncogene 32(36) 4284–4293 https://doi.org/10.1038/onc.2012.433
47. Cochrane DR, Spoelstra NS, and Howe EN, et al (2009) MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents Mol Cancer Ther 8(5) 1055–1066 https://doi.org/10.1158/1535-7163.MCT-08-1046 PMID: 19435871 PMCID: 4573391
48. Wang L, Mezencev R, and Švajdler M, et al (2014) Ectopic over-expression of miR-429 induces mesenchymal-to-epithelial transition (MET) and increased drug sensitivity in metastasizing ovarian cancer cells Gynecol Oncol 134(1) 96–103 [http://www.ncbi.nlm.nih.gov/pubmed/24802724] Date accessed: 26/03/20 https://doi.org/10.1016/j.ygyno.2014.04.055 PMID: 24802724
49. Liu N, Zhou C, and Zhao J, et al (2012) Reversal of paclitaxel resistance in epithelial ovarian carcinoma cells by a muc1 aptamer-let-7i chimera Cancer Invest 30(8) 577–582 [http://www.ncbi.nlm.nih.gov/pubmed/22812695] Date accessed: 13/03/20 https://doi.org/10.3109/07357907.2012.707265 PMID: 22812695
50. Saleh AD, Savage JE, and Cao L, et al (2011) Cellular stress induced alterations in MicroRNA let-7a and let-7b expression are dependent on p53 PLoS One 6(10) e24429 [https://dx.plos.org/10.1371/journal.pone.0024429] Date accessed: 06/04/21 https://doi.org/10.1371/journal.pone.0024429 PMID: 22022355 PMCID: 3191136
51. Tsang WP and Kwok TT (2008) Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3 Apoptosis 13(10) 1215–1222 [http://www.ncbi.nlm.nih.gov/pubmed/18758960] Date accessed: 14/03/20 https://doi.org/10.1007/s10495-008-0256-z PMID: 18758960
52. Sorrentino A, Liu CG, and Addario A, et al (2008) Role of microRNAs in drug-resistant ovarian cancer cells Gynecol Oncol 111(3) 478–486 https://doi.org/10.1016/j.ygyno.2008.08.017 PMID: 18823650
53. Xiao M, Cai J, and Cai L, et al (2020) Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair J Ovarian Res 10(1) 24 [http://www.ncbi.nlm.nih.gov/pubmed/28376831] Date accessed: 12/03/20 https://doi.org/10.1186/s13048-017-0321-8
54. Kuang Y, Cai J, and Li D, et al (2013) Repression of Dicer is associated with invasive phenotype and chemoresistance in ovarian cancer Oncol Lett 5(4) 1149–1154 https://doi.org/10.3892/ol.2013.1158 PMID: 23599754 PMCID: 3629258
55. Boyerinas B, Park SM, and Murmann AE, et al (2012) Let-7 modulates acquired resistance of ovarian cancer to Taxanes via IMP-1-mediated stabilization of multidrug resistance 1 Int J Cancer 130(8) 1787–1797 https://doi.org/10.1002/ijc.26190
56. Meng X, Müller V, and Milde-Langosch K, et al (2016) Circulating cell-free miR-373, miR- 200a, miR-200b and miR-200c in patients with epithelial ovarian cancer Adv Exp Med Biol 924 3–8. https://doi.org/10.1007/978-3-319-42044-8_1
57. Zuberi M, Mir R, and Das J, et al (2015) Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features Clin Transl Oncol 17(10) 779–787 https://doi.org/10.1007/s12094-015-1303-1 PMID: 26063644
58. Wang K, Zhang S, and Weber J, et al (2010) Export of microRNAs and microRNA-protective protein by mammalian cells Nucleic Acids Res 38(20) 7248–7259 https://doi.org/10.1093/nar/gkq601 PMID: 20615901 PMCID: 2978372
59. Choi PW, Bahrampour A, and Ng SK, et al (2020) Characterization of miR-200 family members as blood biomarkers for human and laying hen ovarian cancer Sci Rep 10(1) 20071 [https://pubmed.ncbi.nlm.nih.gov/33208870/] Date accessed: 21/03/21 https://doi.org/10.1038/s41598-020-77068-0 PMID: 33208870 PMCID: 7674435
60. Wang W, Wu LR, and Li C, et al (2019) Five serum microRNAs for detection and predicting of ovarian cancer Eur J Obstet Gynecol Reprod Biol X 3 100017 https://doi.org/10.1016/j.eurox.2019.100017 PMID: 31404211 PMCID: 6687444
61. Zheng H, Zhang L, and Zhao Y, et al (2013) Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer PLoS One 8(11) e77853 [www.plosone.org] Date accessed: 07/05/20 https://doi.org/10.1371/journal.pone.0077853 PMID: 24223734 PMCID: 3815222
62. Meng X, Joosse SA, and Müller V, et al (2015) Diagnostic and prognostic potential of serum MIR-7, MIR-16, MIR-25, MIR-93, MIR-182, MIR-376a and MIR-429 in ovarian cancer patients Br J Cancer 113(9) 1358–1366 [http://www.ncbi.nlm.nih.gov/pubmed/26393886] Date accessed: 21/03/20 https://doi.org/10.1038/bjc.2015.340 PMID: 26393886 PMCID: 4815782
63. Kobayashi M, Salomon C, and Tapia J, et al (2014) Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200 J Transl Med 12(1) 4 https://doi.org/10.1186/1479-5876-12-4 PMID: 24393345 PMCID: 3896684
64. Chung YW, Bae HS, and Song JY, et al (2013) Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient Int J Gyn Can 23(4)