A systematic review and meta-analysis of the prevalence of therapeutic targets in cervical cancer
Maria Guadalupe Patrono1a, Maria Florencia Calvo1b, Juan Victor Ariel Franco2,3c, Virginia Garrote4d and Valeria Vietto2,3e
1Department of Gynecology, Hospital Italiano de Buenos Aires, Gascon 450, Buenos Aires C1181ACH, Argentina
2Family and Community Medicine Division, Hospital Italiano de Buenos Aires, Gascon 450, Buenos Aires C1181ACH, Argentina
3Argentine Cochrane Centre, Instituto Universitario Hospital Italiano, Potosí 4265, Buenos Aires C1199ABB, Argentina
4Central Library, Hospital Italiano de Buenos Aires, Instituto Universitario Hospital Italiano de Buenos Aires,
Tte. J. D. Perón 4190, 1º floor, stair J. C1199ABB, Argentina
ahttps://orcid.org/0000-0002-1309-2114
bhttps://orcid.org/0000-0002-2224-1564
chttps://orcid.org/0000-0003-0411-899X
dhttps://orcid.org/0000-0002-7328-6228
ehttps://orcid.org/0000-0003-4619-9812
Abstract
Cervical Cancer (CC) is a significantly prevalent disease in developing countries. Currently, targeted therapies are not a primary standard of care in CC. This information could be crucial for developing directed therapies and patient screening for biomarkers that would allow personalised treatment of CC. This systematic review aimed to estimate the prevalence of potential therapeutic targets such as the epidermal growth factor receptor (EGFR) and the PI3K/Akt/mTOR and Ras/Raf/MAPK pathways in patients with CC, identified through genomic and non-genomic testing. Studies were identified through an ad-hoc search strategy from the available on MEDLINE (Ovid), CENTRAL, LILACS, SCOPUS, through the Clinical Trial registry on Clinicaltrials.gov, International Clinical Trials Registry Platform, RENIS (Argentine National Registry of Health Research) and grey literature sources. We included 74 studies which represented a total pool of 7,862 participants. Forty-five studies informed mutations of EGFR, with a combined positivity rate of 53% (95%CI: 45%–60%; I2 = 95%). Twenty studies informed the presence of mutations in PIK3CA with a combined positivity rate of 30% (95%CI: 21%–39%; I2 = 96%). Twenty-three studies reported a mutation in Ras, with a combined positivity rate of 14% (95%CI: 8%–21%; I2 = 95%). Raf mutations were informed in six studies. Six studies informed the presence of Akt mutations, two studies informed mTOR mutations and only one study reported mutations of MAPK. The most frequently described therapeutic targets were EGFR, and the PIK3CA and Ras pathways, though inconsistency in positivity rates was significant. Our study did not allow the identification of any specific clinical characteristics that might explain the observed heterogeneity. Despite the overall good quality of the included studies, the applicability of these results to patients’ general population with CC is still unclear.
Keywords: cervical cancer, cervical neoplasia, mutation, prevalence, therapeutic targets
Correspondence to: Maria Guadalupe Patrono
Email: maria.patrono@hospitalitaliano.org.ar
Published: 09/03/2021
Received: 25/10/2020
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Cervical-uterine cancer is the second most diagnosed cancer in women in developing countries. About 4,400 new cases are diagnosed in Argentina, and approximately 1,800 women die from this disease each year. Globally, in 2018, 570,000 women were diagnosed with cervical cancer (CC), and 311,000 died from this cause, constituting the fourth in frequency among women and the fourth as a cause of death. Approximately 84% of all CCs and 88% of all deaths caused by this cancer occurred in lower resource countries [1–3].
Infection by human papillomavirus (HPV) has been clearly established as a necessary cofactor for the development of CC. The use of cervical cytology, known as Papanicolaou testing, as well the addition of HPV co-testing has significantly improved the detection and treatment of precursor and preinvasive cervical lesions, allowing to identify patients who are at greater risk of developing CC [1].
Standard treatment for women with early-stage CC (International Federation of Gynecology and Obstetrics (FIGO) stages IA and IB) consists of radical hysterectomy or concurrent radiotherapy/chemotherapy treatment. Survival rates are similar with either treatment modality. The choice of therapy usually depends on either treatment’s adverse event profile, the presence or absence of specific prognostic factors, patient or physician preference and access to the different therapeutic options [2]. In patients presenting with advanced stages of CC, both neoadjuvant chemotherapy and concurrent radiotherapy/chemotherapy are suitable alternatives. Still, neoadjuvant chemotherapy before surgery or chemoradiation is associated with inferior outcomes compared to concurrent chemoradiation [2, 4, 5].
Nonetheless, therapeutic options have been evolving continuously, along with the discovery and development of targeted therapies. Genetic profiling of tumours could predict sensitivity or resistance to these novel treatments, facilitating a personalised approach for each patient. Identifying specific genetic tumour alterations or the presence of distinct targets can guide treatment with molecular agents and provide prognostic information about the disease [2, 6, 7].
In 1997, the Food and Drug Administration of the United States of America approved the first targeted therapy, rituximab (Rituxan®), to treat patients with Non-Hodgkin B-cell lymphoma who had progressed to other treatments. Currently, numerous tumours such as breast, ovarian, liver, prostate, lung cancer and melanoma have targeted therapies, which have proven beneficial in improving disease-free survival and overall survival in some cases. Regarding CC, one of the first studies investigating the use of the specific anti-PDLI-1 monoclonal antibody Pembrolizumab in patients with recurrent disease was published in 2019 [8].
However, targeted therapies have not yet been established as a primary treatment standard in CC. One of the main limitations for the development of directed therapy is that the prevalence of mutations or specific molecular targets in CC is still unclear, thus making it impossible to screen and identify patients who may be suitable candidates. The characterisation of the most frequent mutations in patients with CC is necessary. It will allow the development of detection kits for specific markers with prognostic and predictive potential, which may be used in clinical practice to guide and personalise oncologic treatment for each patient.
Well-established markers as therapeutic options in many tumours include the epidermal growth factor receptor (EGFR, ErbB), phosphatidylinositol 3-kinase (PI3K)/Akt/mTOR and Ras/Raf/MAPK [9–11]. The epidermal growth factor receptor (EGFR, ErbB) belongs to the superfamily of plasma membrane-localised receptors with intrinsic tyrosine kinase activity, activated by the epidermal growth factor (EGF) and the tumour growth factor alpha. EGFR is activated by dimerisation, which depends on ligand binding, although it can also occur when there is overexpression and structural alterations of the receptor. Signalling through the EGFR is crucial in embryonic development: in epithelial development, cell proliferation and organogenesis. Uncontrolled EGFR activity (by overexpression or structural abnormalities in the receptor or its ligands) has been implicated in many aspects of tumour growth, including the promotion of cell proliferation, angiogenesis, invasion, metastasis and survival. When an extracellular ligand binds to EGFR, activation of its tyrosine kinase and phosphorylation of tyrosine residues occur. The best known and best-characterised signalling pathway initiated by activated EGFR is the Ras/MAPK pathway, which appears to be essential for EGF-mediated cell proliferation. Another important pathway is the PI3K, and the PI3K/Akt/mTOR cascade, which results in increased protein transcription, synthesis and proliferation. The PIK3CA gene encodes the p110 alpha catalytic subunit of PI3K, which is often mutated in tumoural cells [12–15].
This systematic review aimed to estimate the prevalence of potential and established therapeutic targets in patients with CC. We considered the following markers: EGFR, and both PI3K/Akt/mTOR and Ras/Raf/MAPK intracellular pathways.
Materials and methods
A systematic review was performed, according to the methodologic guidelines of the Manual for Systematic Reviews of Observational Studies of the Joanna Briggs Institute [16, 17] and the checklist for Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) [18]. The study protocol was approved by the Internal Review Board of the Hospital Italiano de Buenos Aires (Ethics Committee in Research Protocols, CEPI) and was registered in the PROSPERO database of systematic reviews (ID CRD42017067538).
Search methods
The search was performed using the following electronic databases: MEDLINE (Ovid), CENTRAL, LILACS, SCOPUS and the registry of Clinical Trials Clinicaltrials.gov, the International Clinical Trials Registry Platform (ICTRP) and RENIS until September 2019. The search strategy was designed and executed by a librarian specialised in systematic reviewing. The full search strategy, which was initially developed for MEDLINE (Ovid) and later adjusted for the other search engines, is listed in Appendix A. Searches were also performed in grey literature sources, through Google Scholar and OpenGrey. The references of the included studies were revised, and the summaries and abstracts of the most relevant speciality meetings and conferences of the past 5 years were manually explored: Annual Meeting of the American Society of Gynecologic Oncologist, Annual Meeting of the International Gynecologic Cancer Society, Annual Meeting of European Society of Medical Oncology and Annual Meeting of the American Society of Clinical Oncology.
Study selection criteria
For this systematic review, the inclusion of primary studies was considered for randomised and quasi-randomised controlled trials, observational cohort studies and cross-sectional studies, including patients of any age, diagnosed with primary, non-recurrent squamous, adenosquamous or adenocarcinoma CC, in whom expression of one or more therapeutic or potential therapeutic targets were explored. Surface protein expression and intracellular genomic expression and amplification were both considered positive results. The investigated targets were the following: EGFR and both PI3K/Akt/mTOR and Ras/Raf/MAPK pathways, with or without HPV virus typification. Studies performed in commercial cell cultures, patients with CC recurrence and those with unavailable or incomplete data to estimate the proportion of participants with a positive test were excluded. Inclusion was limited to English or Spanish reports.
Study selection procedure
Study screening and selection were performed by two independent reviewers (MGP and MFC), using the online platform COVIDENCE. The first stage involved evaluating the titles and abstracts of all records retrieved by the search strategy. The second stage consisted of the full-text revision. The full texts considered relevant after initial screening were recovered and analysed according to the research question. Reviewer discrepancies were resolved by consensus or by a third reviewer (VV).
Data extraction
Data extraction was performed by completing an ad-hoc standardised electronic form (Google Form®), after independent proofing by two reviewers (MGP and MFC). Discrepancies were resolved by consensus or by a third reviewer (VV).
The following data were collected: study design and aim, language, year of publication, inclusion and exclusion criteria and characteristics of the study population, which included sample size, age at diagnosis, tumour histology (squamous, adenosquamous or adenocarcinoma) stage of disease at diagnosis according to the International Federation of Gynecology and Obstetrics staging criteria and type of treatment. Additional variables that were documented were tumour size, parametrial invasion, depth of myocervical invasion, lymphovascular space invasion, presence of pelvic or paraaortic lymph node metastases, adjuvant treatment and HPV typification.
The primary outcome was to describe the prevalence of targets or potential targets, defined as positive expression (either genomic or nongenomic) of one or more of the following markers, independent of histologic tumour subtype or stage: EGFR and both PI3K/Akt/mTOR and Ras/Raf/MAPK pathways.
Strategies for data synthesis
Statistical analysis was performed using STATA® software (StataCorp, College Station, TX). The total percentage of variation among studies due to heterogeneity was assessed using the I² test. Low heterogeneity was defined for values between 0% and 40%, moderate for 30% through 60%, substantial between 50% and 90% and considerable heterogeneity was established between 75% and 100%. Since high heterogeneity was expected, proportions were combined through meta-analysis using a random-effects model [19]. Combined estimations were calculated with 95% confidence intervals and were represented through forest plot. We attempted to explore heterogeneity by performing prespecified subgroup analysis, for both histologic subtype and detection method (genomic versus nongenomic). However, the data were insufficient for performing these analyses or did not gather enough power to detect differences.
Sensibility analysis
Performance of sensitivity analysis was intended at the study onset, in order to explore the risk of possible selection and information bias influencing the proportion of positivity of the detection tests for the therapeutic targets. These analyses were not performed since none of the included studies was considered to be at high risk of bias in either domain.
Assessment for publication bias
Funnel plots were employed to evaluate small studies’ effect whenever at least ten studies were available for a specific outcome.
Assessment of study quality and bias
The methodological quality of the included studies was independently assessed by each reviewer, by using the ‘Critical Appraisal by the Joanna Briggs Institute for prevalence studies’ tool, adapted ad-hoc for this review [20, 21]. Through this structured and standardised instrument consisting of seven specific questions, key aspects of the study population, setting and design were explored, such as detailed population description, adequate, appropriate and representative sampling, standardised measurement of outcomes and statistical aspects, including appropriate sample size and accurate use of statistical methods for data extraction (Appendix C).
Results
Study selection and inclusion
The study selection process is summarised in Figure 1. After executing the search strategy, which is described in Appendix A, a total of 3,935 studies were retrieved. Among these, 1,256 were automatically detected as duplicates by the reference management software and excluded. Manual search did not find additional studies.
After the initial title and abstract screening process of all the initially recovered articles, 167 studies were selected for full-text review. After this stage, 74 studies that gathered the established inclusion criteria for our revision were included. The complete list of the 93 excluded studies can be found in Appendix B. The studies are grouped according to the reason for exclusion, which included impossibility of access to the full text (in all cases, direct email contact with the author was attempted but not achieved), inappropriate study design (narrative or systematic reviews), inappropriate study population (e.g. mutations described in cell cultures or cell lines or tumour histologies other than the prespecified types) and those that did not allow confidable data extraction.

Figure 1. PRISMA flow diagram.
Characteristics of the included studies
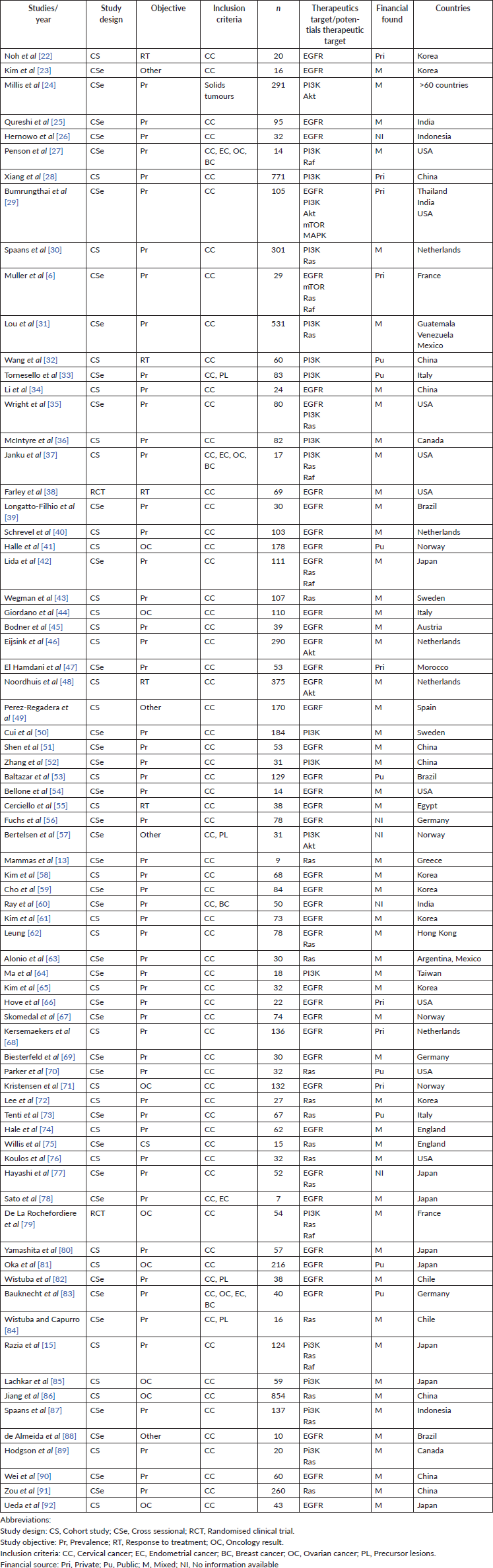
The 74 included (Table 1) studies were published between 1989 and 2019. Thirty-nine of the studies had a cross-sectional design (52.7%), 33 (44.6%) were observational cohort studies and 2 (2.7%) were randomised clinical trials, which resulted in a pooled total of 7,862 patients (median: 106, interquartile range: 30–110, range: 7–854). Forty-five studies (60.8%) specified patient age, with a resulting mean of 49 years (standard deviation: 5.73, range: 39–69).
With regard to the primary outcome, 57 studies (77%) assessed the prevalence of or proportion of specific mutations, 5 (6.7%) evaluated response to treatment, 8 (10.8%) focused on patient outcome and the remaining 4 had other non-clinically applicable aims (5.4%).
English was the preferred language for publication in 71 articles (96%). The remaining 3 were in Spanish. Only 4 studies (5.4%) were multicentric, led by investigators from different countries, while 70 were performed in single countries (94.6%). The study population was grossly represented by patients with CC (64 studies; 86.4%), but four studies (5.4%) also included patients with precursor lesions and five (6.7%) involved patients with other gynaecologic cancers (including breast), and one study included other solid tumours (pancreas and colon, among others).
Tumour histology data was not extractable in seven studies (9.4%), representing 752 patients. The remaining studies pooled a total of 6,886 (87.6%) patients, among which 5,377 (68.4%) had squamous histology, 394 (5%) had adenosquamous subtypes and 1,115 (14.2%) were adenocarcinomas.
Thirty-seven studies (50%) detailed the type of treatment received by the study population, whether it was concomitant chemo/radiation therapy, neoadjuvant chemotherapy or primary surgery. Forty-two studies (56.7%) included data regarding patient outcome and prognosis, such as disease-free survival or overall survival. Twenty-two studies (29.7%) also stated the study population’s HPV status and several specified subtypes.
In six of the included studies (8.1%), the authors declared conflicts of interest. Twenty-six declared having no disclosures whatsoever (35.1%). In the remaining studies, this information was unavailable. Regarding funding and grant support, 10.2% received public funding, 10.2% received private grants and 71.6% had a combined source, through grants from universities, laboratories or scholarships. Funding was unspecified in five studies (9.5%).
Table 1. Characteristics of the included studies.
Methodological quality of the included studies
In all of the included studies, the sample population was found to adequately represent the target population (criteria 1), which means that the patients with CC included, complied with the selected subtypes’ prerequisite (squamous, adenosquamous or adenocarcinoma). The majority of the studies (97%) recruited the study population adequately (criteria 2), so the risk of selection bias was considered low.
Less than 6% of the included studies reported accurate sample size calculation (criteria 3), while in most cases (94%) this parameter was not assessable, given that the method for the definition of sample size was either not informed or was not considered applicable to the primary aim of the study.
The majority of the studies (77%) accurately described the study population (criteria 4). It was partially described in 13.5% and 9.5% did not provide any information regarding the characteristics of the population. All of the included studies (100%) performed data analysis with adequate coverage of the population and used one or more of the prespecified standard methods for measuring the variable of interest (100%), which is why the risks of data loss and information bias were considered low (criteria 5 and 6).
In the vast majority of the included studies, the described statistical analysis method was adequate for the study aim (93%). Statistical analysis was unclear or uninformed in 5.4% and in one case, was considered inadequate by both reviewers (criteria 7) (Figure 2).
Revision findings
The following is a description of the results for each specific therapeutic or potential therapeutic target.
EGFR
Forty-five studies (61%) representing a combined population of 3,605 patients informed the presence of this mutation [6, 22, 23, 25, 26, 29, 34, 35, 38–42, 44–49, 51, 53–56, 58–62, 65–69, 71, 74, 77, 78, 80–83, 88, 90, 92]. The combined positivity rate was 53% (95%CI: 45%–60%; I2 = 95%; Figure 1, Appendix D). Nonetheless, considering the substantial heterogeneity, the positivity rate varied from 4% to 100%. Potential explanations for this heterogeneity were explored, but no differences relative to histological subtype, testing method (genomic versus non-genomic) or any other clinical characteristics were established (Figure 2, Appendix D).
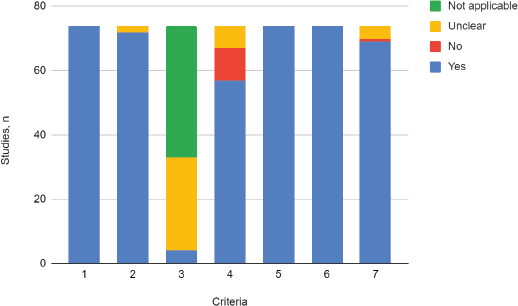
Figure 2. Critical appraisal. Adaptation of the criteria of the ‘Joanna Briggs Institute Critical Appraisal Checklist for Prevalence Studies’.
1. Was the sample frame appropriate to address the target population?
2. Were study participants recruited in an appropriate way?
3. Was the sample size adequate?
4. Were the study subjects and setting described in detail?
5. Was data analysis conducted with sufficient coverage of the identified sample?
6. Was the condition measured in a standard, reliable way for the variable of interest in all cases?
7. Was there appropriate statistical analysis?
PIK3CA
Twenty studies (27%) pooling 2,993 participants informed the presence of this mutation [15, 24, 27–33, 35–37, 50, 52, 57, 64, 79, 85, 87, 89]. The combined positivity rate was 30% (95%CI: 21%–39%; I2 = 96%; Figure 4, Appendix D). Considering the substantial heterogeneity, the positivity rate varied between 6% and 81%. Possible explanations for this heterogeneity were explored, but no differences for histologic subtypes were found (Figure 5, Appendix D). The influence of the type of test used was not tested due to the insufficient number of studies in each subgroup, nor the observed inconsistency could be attributed to any other clinical condition.
Ras
Twenty-three studies (31%) with 2,983 participants reported this mutation [6, 13, 15, 30, 31, 35, 37, 42, 43, 62, 63, 70, 72, 73, 75–77, 79, 84, 86, 87, 89, 91]. The combined positivity rate was 14% (95%CI: 8%–21%; I2 = 95%, Figure 7, Appendix D); however, considering the substantial, heterogeneity, the positivity rate ranged between 0% and 82%. When possible explanations for this heterogeneity were explored, higher positivity rates for this marker were found in the squamous subtype than in adenocarcinomas (pooled estimates 6% versus 30%, p < 0.01; Figure 8, Appendix D). Also, studies that used non-genomic testing described higher positivity rates than those that employed genomic tests (pooled estimates 13% versus 39%, p < 0.01; Figure 9, Appendix D). No additional clinical characteristics that could account for the observed heterogeneity were identified.
Akt
Six studies (8%) gathering 1,106 participants informed the presence of this mutation [24, 27, 29, 46, 48, 57]. A quantitative synthesis of all studies was not performed since two different groups were clearly identified; four studies with 970 participants informed very low positivity rates (globally 2%; 95%CI: 0%–5%) and two studies including 136 participants reported high positivity rates (total 88%; 95%CI: 82%–93%) (Figure 11, Appendix D). No clinical characteristics that could explain this inconsistency were identified.
Raf
Raf mutations were informed in six studies (8%) with a total of 349 participants [6, 15, 27, 37, 42, 79]. The pooled estimate for positivity rate was 1% (95%CI: 0%–4%; I2 = 38%; Figure 12, Appendix D). No clinical characteristics that could explain this inconsistency were identified.
mTOR
Two studies (2.7%) reported this mutation with different results [6, 29]. Muller et al [6] informed that the positivity rate was 3% (1/29 study subjects), while Bumrungthai et al [29] reported a 61% rate (64/105 subjects). No clinical characteristics that could explain this inconsistency were identified.
MAPK
Only one study with 105 participants (1.3%) [29] reported this mutation with a positivity rate of 68% (n = 71).
Risk of publication bias
We detected significant asymmetry in the funnel plots, which could indicate the presence of publication bias for the mutation positivity proportions of EGFR and PIK3CA, while the asymmetry was less evident for Ras (Figures 3, 6 and 10, Appendix D). Given that the plots can only be represented in outcomes that group at least ten studies, we could not assess the effect of the smaller studies for the remaining markers.
Discussion
To the authors’ knowledge, this is the first systematic review evaluating the prevalence or proportion of potential and established therapeutic targets in CC. The identification of these markers may allow the development of future research with crucial clinical applicability, such as the screening for specific predictive factors at the onset of treatment, as is already being performed in other tumours. This may ultimately lead to the improvement of clinical outcomes in patients diagnosed with this disease through the use of precision medicine. One example of the benefit of using target therapy to improve outcomes in a different clinical setting is the higher progression-free survival and quality of life by targeting BRAF mutations in patients with metastatic colon cancer. This practice, which currently constitutes a standard of care, allows the identification of patients who will benefit from monoclonal antibody therapy targeting EGFR (cetuximab, panitumumab, among others) [9].
According to our findings, the most frequently explored markers in patients with CC are the EGFR and both PIK3CA and Ras pathways. We were unable to identify clinical characteristics that might justify the substantial inconsistencies observed among these studies for most markers. However, in the case of Ras, we found that the histologic subtype and the nature of the detection test employed could affect the positivity rate of this target (higher in squamous carcinoma and with the use of non-genomic testing). Because of this, the pooled estimates for the prevalence of therapeutic targets were considered to be low-certainty evidence, and the value ranges extracted from primary studies should be highlighted as the main features of this systematic review.
We considered both surface expression and intracellular gene amplification positive results since research is underway in both tumour genome and surface markers that may prove useful for the identification of clinically significant alterations that will probably become relevant in personalising patient treatment and care.
Among the limitations of our study, we can describe the limitations due to the unavailability of language translation resources, which narrowed our options to including only full texts published in English or Spanish. However, we only excluded eight studies for this reason, which is why we consider that the potential impact of this limitation is probably insignificant.
On the other hand, the methodological quality appraisal and the evaluation of the risk of bias in the collected evidence may be somewhat limited. One of the methodological difficulties of systematic reviews of prevalence resides in the diversity of instruments for the appraisal of primary studies. This is in part because there is a lack of consensus regarding which domains need to be evaluated in this study type, and there is no universally accepted assessment tool. For this review, we decided to create an ad-hoc adaptation of the ten criteria of the ‘Joanna Briggs Institute Critical Appraisal Checklist for Prevalence Studies’, since we considered it to be the most applicable to our study aim. This structured assessment tool was developed for any clinical condition based on the systematic review of previous instruments. Its validity, applicability and acceptability have been appreciated by experts and may be currently considered the most complete appraisal tool according to a recent review [93].
In general, the quality of the studies included in this revision was adequate. One of the greatest weaknesses that we identified through this instrument was the lack of sample size estimation, which turned out to be a frequent finding in our study pool. Most of the studies reported results in terms of proportions and had not been designed to determine the prevalence of the evaluated markers within the universe of patients with CC. Also, the primary studies did not apply sampling techniques that could guarantee the representativity of the analysed specimens, allowing the generalisation of their results.
The development of directed therapies requires the identification of adequate markers. These are specific targets or checkpoints that play a crucial role in malignant cell proliferation and growth. The assessment of the prognostic and predictive clinical value of these markers lies without the scope of this systematic review and may be an appropriate subject of further research. However, the use of these treatments in other tumours has proven a significant impact on patient outcomes and quality of life and hopefully, this may be soon applicable to CC patients as well.
From a health economics standpoint, it is important also to consider the costs associated with the systematic detection of the targets to implement precision therapies. This will require the performance of economic assessments and the development of clinical trials with patient-centred outcomes that will assess the financial burden of the specific interventions that may result beneficial in each particular context.
Conclusion
This systematic review found that the most frequently described therapeutic targets were EGFR and the PIK3CA and Ras pathways. Nonetheless, the reported estimates of positivity prevalence for these markers are inconsistent. Our study did not allow the identification of any specific clinical characteristic that might explain the observed heterogeneity, except for the Ras pathways, which showed higher rates of positivity in squamous histologic subtypes and when non-genomic assays were used for testing. Despite the overall high quality of the included studies, the applicability of these results to patients’ general population with CC is still unclear.
Authors’ contributions
Concept and design: Patrono MG, Calvo MF, Franco JV, Vietto V.
Acquisition, analysis or interpretation of data: Patrono MG, Calvo MF, Franco JV, Garrote V, Vietto V.
Drafting of the manuscript: Patrono MG, Calvo MF, Vietto V.
Critical revision of the manuscript for important intellectual content: Patrono MG, Calvo MF, Franco JV, Garrote V, Vietto V.
Statistical analysis: Franco JV, Vietto V.
Supervision: Vietto V.
Conflicts of interest
The authors have no conflicts of interest to declare.
Funding statement
None.
References
1. Arrossi S, Thouyaret L and Luis P (2015) Recomendaciones para el tamizaje, seguimiento y tratamiento de mujeres en el marco de programas de tamizaje basados en el test de VPH ACTUALIZACIÓN
2. Dueñas-González A and Campbell S (2016) Global strategies for the treatment of early-stage and advanced cervical cancer Curr Opin Obstet Gynecol 28(1) 11–17 https://doi.org/10.1097/GCO.0000000000000234
3. Arbyn M, Weiderpass E, and Bruni L, et al (2020) Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis Lancet Glob Health 8 e191–e203 https://doi.org/10.1016/S2214-109X(19)30482-6 PMCID: 7025157
4. Chuang LT, Temin S, and Camacho R, et al (2016) Management and care of women with invasive cervical cancer: American Society of Clinical Oncology Resource-Stratified Clinical Practice Guideline J Glob Oncol 2 311–340 https://doi.org/10.1200/JGO.2016.003954 PMID: 28717717 PMCID: 5493265
5. Gupta S, Maheshwari A, and Parab P, et al (2018) Neoadjuvant chemotherapy followed by radical surgery versus concomitant chemotherapy and radiotherapy in patients with stage IB2, IIA, or IIB squamous cervical cancer: a randomized controlled trial J Clin Oncol 36 1548–1555 https://doi.org/10.1200/JCO.2017.75.9985 PMID: 29432076
6. Muller E, Brault B, and Holmes A, et al (2015) Genetic profiles of cervical tumors by high-throughput sequencing for personalized medical care Cancer Med 4 1484–1493 https://doi.org/10.1002/cam4.492 PMID: 26155992 PMCID: 4618619
7. Markman B, Atzori F, and Pérez-García J, et al (2009) Status of PI3K inhibition and biomarker development in cancer therapeutics Ann Oncol 21(4) 683–691 https://doi.org/10.1093/annonc/mdp347 PMID: 19713247
8. Chung HC, Ros W, and Delord JP, et al (2019) Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: results from the phase II KEYNOTE-158 study J Clin Oncol 37 1470–1478 https://doi.org/10.1200/JCO.18.01265 PMID: 30943124
9. National Comprehensive Cancer Network Colon Cancer (Version 4.2020).
10. National Comprehensive Cancer Network Kidney Cancer (Version 2.2021).
11. National Comprehensive Cancer Network Melanoma Cutaneous (Version 1.2021).
12. Vivanco I and Sawyers CL (2002) The phosphatidylinositol 3-Kinase AKT pathway in human cancer Nat Rev Cancer 2 489–501 https://doi.org/10.1038/nrc839 PMID: 12094235
13. Mammas IN, Zafiropoulos A, and Koumantakis E, et al (2004) Transcriptional activation of H- and N-ras oncogenes in human cervical cancer Gynecol Oncol 92 941–948 https://doi.org/10.1016/j.ygyno.2003.11.040 PMID: 14984964
14. Manzo-Merino J, Contreras-Paredes A, and Vázquez-Ulloa E, et al (2014) The role of signaling pathways in cervical cancer and molecular therapeutic targets Arch Med Res 45 525–539 https://doi.org/10.1016/j.arcmed.2014.10.008 PMID: 25450584
15. Razia S, Nakayama K, and Nakamura K, et al (2019) Clinicopathological and biological Analysis of PIK3CA mutation and amplification in cervical carcinomas Exp Ther Med 18 2278–2284 PMID: 31410178 PMCID: 6676184
16. Munn Z, MClinSc SM, and Lisy K, et al (2015) Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data Int J Evid Based Healthc 13 147–153 https://doi.org/10.1097/XEB.0000000000000054 PMID: 26317388
17. Aromataris EMZ (2020) JBI Manual for Evidence Synthesis (Adelaide: JBI) [https://synthesismanual.jbi.global]
18. Moher D, Liberati A, and Tetzlaff J, et al (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement BMJ 339 b2535 https://doi.org/10.1136/bmj.b2535 PMID: 19622551 PMCID: 2714657
19. Nyaga VN, Arbyn M, and Aerts M (2014) Metaprop: a Stata command to perform meta-analysis of binomial data Arch Public Health 72 39 https://doi.org/10.1186/2049-3258-72-39 PMID: 25810908 PMCID: 4373114
20. Munn Z, Moola S, and Riitano D, et al (2014) The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence Int J Health Policy Manag 3 123–128 https://doi.org/10.15171/ijhpm.2014.71 PMID: 25197676 PMCID: 4154549
21. Harder T (2014) Some notes on critical appraisal of prevalence studies: Comment on: “The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence.” Int J Health Policy Manag 3 289–290 https://doi.org/10.15171/ijhpm.2014.99 PMID: 25337603 PMCID: 4204748
22. Noh JM, Park W, and Huh SJ, et al (2009) Correlation between tumor volume response to radiotherapy and expression of biological markers in patients with cervical squamous cell carcinoma J Gynecol Oncol 20 215–220 https://doi.org/10.3802/jgo.2009.20.4.215 PMID: 20041097 PMCID: 2799019
23. Kim SC, Park H-M, and Lee S-N, et al (2004) Expression of epidermal growth factor receptor in cervical tissue and serum in patients with cervical neoplasia J Low Genit Tract Dis 8 292–297 https://doi.org/10.1097/00128360-200410000-00006
24. Millis SZ, Ikeda S, and Reddy S, et al (2016) Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors JAMA Oncol 2 1565–1573 https://doi.org/10.1001/jamaoncol.2016.0891 PMID: 27388585
25. Qureshi R, Arora H, and Biswas S, et al (2016) Mutation analysis of EGFR and its correlation with the HPV in Indian cervical cancer patients Tumor Biol 37 9089–9098 https://doi.org/10.1007/s13277-016-4789-4
26. Hernowo BS, Suryanti S, and Wibisono F (2016) Correlation between EGFR expression and radiosensitivity in cervical adenocarcinoma cases Asian Pac J Cancer Prev 17 2535–2537 PMID: 27268625
27. Penson RT, Sales E, and Sullivan L, et al (2016) A SNaPshot of potentially personalized care: molecular diagnostics in gynecologic cancer Gynecol Oncol 141 108–112 https://doi.org/10.1016/j.ygyno.2016.02.032 PMID: 27016236
28. Xiang L, Jiang W, and Li J, et al (2015) PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer Sci Rep 5 14035. https://doi.org/10.1038/srep14035 PMID: 26358014 PMCID: 4566086
29. Bumrungthai S, Munjal K, and Nandekar S, et al (2015) Epidermal growth factor receptor pathway mutation and expression profiles in cervical squamous cell carcinoma: therapeutic implications J Transl Med 13 244 https://doi.org/10.1186/s12967-015-0611-0 PMID: 26209091 PMCID: 4513684
30. Spaans VM, Trietsch MD, and Peters AAW, et al (2015) Precise classification of cervical carcinomas combined with somatic mutation profiling contributes to predicting disease outcome PLoS One 10 https://doi.org/10.1371/journal.pone.0133670 PMID: 26197069 PMCID: 4510875
31. Lou H, Villagran G, and Boland JF, et al (2015) Genome analysis of Latin American cervical cancer: frequent activation of the PIK3CA pathway Clin Cancer Res 21 5360–5370 https://doi.org/10.1158/1078-0432.CCR-14-1837 PMID: 26080840 PMCID: 4668220
32. Wang J, Chai Y-L, and Wang T, et al (2015) Genetic alterations of PIK3CA and tumor response in patients with locally advanced cervical squamous cell carcinoma treated with cisplatin-based concurrent chemoradiotherapy Exp Mol Pathol 98 407–410 https://doi.org/10.1016/j.yexmp.2015.03.014 PMID: 25773678
33. Tornesello ML, Annunziata C, and Buonaguro L, et al (2014) TP53 and PIK3CA gene mutations in adenocarcinoma, squamous cell carcinoma and high-grade intraepithelial neoplasia of the cervix J Transl Med 12 255 https://doi.org/10.1186/s12967-014-0255-5 PMID: 25220666 PMCID: 4174264
34. Li Q, Tang Y, and Cheng X, et al (2014) EGFR protein expression and gene amplification in squamous intraepithelial lesions and squamous cell carcinomas of the cervix Int J Clin Exp Pathol 7 733–741 PMID: 24551297 PMCID: 3925921
35. Wright AA, Howitt BE, and Myers AP, et al (2013) Oncogenic mutations in cervical cancer: Genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix Cancer 119 3776–3783 https://doi.org/10.1002/cncr.28288 PMID: 24037752 PMCID: 3972000
36. McIntyre JB, Wu JS, and Craighead PS, et al (2013) PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy Gynecol Oncol 128 409–414 https://doi.org/10.1016/j.ygyno.2012.12.019
37. Janku F, Wheler JJ, and Westin SN, et al (2012) PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations J Clin Oncol 30 777–782 https://doi.org/10.1200/JCO.2011.36.1196 PMID: 22271473 PMCID: 3295566
38. Farley J, Sill MW, and Birrer M, et al (2011) Phase II study of cisplatin plus cetuximab in advanced, recurrent, and previously treated cancers of the cervix and evaluation of epidermal growth factor receptor immunohistochemical expression: a Gynecologic Oncology Group study Gynecol Oncol 121 303–308 https://doi.org/10.1016/j.ygyno.2011.01.030 PMID: 21329967 PMCID: 3081894
39. Longatto-Filho A, Pinheiro C, and Martinho O, et al (2009) Molecular characterization of EGFR, PDGFRA and VEGFR2 in cervical adenosquamous carcinoma BMC Cancer 9 https://doi.org/10.1186/1471-2407-9-212 PMID: 19563658 PMCID: 2711112
40. Schrevel M, Karim R, and Ter Haar NT, et al (2012) CXCR7 expression is associated with disease-free and disease-specific survival in cervical cancer patients Br J Cancer 106 1520–1525 https://doi.org/10.1038/bjc.2012.110 PMID: 22531719 PMCID: 3341866
41. Halle C, Lando M, and Svendsrud DH, et al (2011) Membranous expression of ectodomain isoforms of the epidermal growth factor receptor predicts outcome after chemoradiotherapy of lymph node-negative cervical cancer Clin Cancer Res 17 5501–5512 https://doi.org/10.1158/1078-0432.CCR-11-0297 PMID: 21737508
42. Iida K, Nakayama K, and Rahman MT, et al (2011) EGFR gene amplification is related to adverse clinical outcomes in cervical squamous cell carcinoma, making the EGFR pathway a novel therapeutic target Br J Cancer 105 420–427 https://doi.org/10.1038/bjc.2011.222 PMID: 21730982 PMCID: 3172895
43. Wegman P, Ahlin C, and Sorbe B (2011) Genetic alterations in the K-Ras gene influence the prognosis in patients with cervical cancer treated by radiotherapy Int J Gynecol Cancer 21 86–91 https://doi.org/10.1097/IGC.0b013e3182049924 PMID: 21330833
44. Giordano G, D’Adda T, and Dal Bello B, et al (2011) Clinicopathologic implications of the epidermal growth factor receptor, cyclooxygenase 2 expression, and human papillomavirus status in squamous cell carcinoma of the uterine cervix in the elderly Int J Gynecol Cancer 21 337–348 https://doi.org/10.1097/IGC.0b013e31820864b7 PMID: 21270615
45. Bodner K, Laubichler P, and Kimberger O, et al (2011) Expression of p16 protein and epidermal growth factor receptor in patients with adenocarcinoma of the uterine cervix: an immunohistochemical analysis Arch Gynecol Obstet 283 611–616 https://doi.org/10.1007/s00404-010-1464-7
46. Eijsink JJH, Noordhuis MG, and Ten Hoor KA, et al (2010) The epidermal growth factor receptor pathway in relation to pelvic lymph node metastasis and survival in early-stage cervical cancer Hum Pathol 41 1735–1741 https://doi.org/10.1016/j.humpath.2010.04.017 PMID: 21078436
47. El Hamdani W, Amrani M, and Attaleb M, et al (2010) EGFR, p16INK4a and E-cadherin immuno- histochemistry and EGFR point mutations analyses in invasive cervical cancer specimens from Moroccan women Cell Mol Biol 56 OL1373–OL1384
48. Noordhuis MG, Eijsink JJH, and Ten Hoor KA, et al (2009) Expression of epidermal growth factor receptor (EGFR) and activated EGFR predict poor response to (Chemo)radiation and survival in cervical cancer Clin Cancer Res 15 7389–7397 https://doi.org/10.1158/1078-0432.CCR-09-1149 PMID: 19920104
49. Pérez-Regadera J, Sánchez-Muñoz A, and De-La-Cruz J, et al (2009) Negative prognostic impact of the coexpression of epidermal growth factor receptor and c-erbB-2 in locally advanced cervical cancer Oncology 76 133–141 https://doi.org/10.1159/000195539 PMID: 19174612
50. Cui B, Zheng B, and Zhang X, et al (2009) Mutation of PIK3CA: Possible risk factor for cervical carcinogenesis in older women Int J Oncol 34 409–416 PMID: 19148475
51. Shen L, Shui Y, and Wang X, et al (2008) EGFR and HER2 expression in primary cervical cancers and corresponding lymph node metastases: implications for targeted radiotherapy BMC Cancer 8 https://doi.org/10.1186/1471-2407-8-232
52. Zhang X-Y, Zhang H-Y, and Zhang P-N, et al (2008) Elevated phosphatidylinositol 3-kinase activation and its clinicopathological significance in cervical cancer Eur J Obstet Gynecol Reprod Biol 139 237–244 https://doi.org/10.1016/j.ejogrb.2007.12.021 PMID: 18395956
53. Baltazar F, Filho AL, and Pinheiro C, et al (2007) Cyclooxygenase-2 and epidermal growth factor receptor expressions in different histological subtypes of cervical carcinomas Int J Gynecol Pathol 26 235–241 https://doi.org/10.1097/pgp.0b013e31802f1996 PMID: 17581404
54. Bellone S, Frera G, and Landolfi G, et al (2007) Overexpression of epidermal growth factor type-1 receptor (EGF-R1) in cervical cancer: Implications for Cetuximab-mediated therapy in recurrent/metastatic disease Gynecol Oncol 106 513–520 https://doi.org/10.1016/j.ygyno.2007.04.028 PMID: 17540437
55. Cerciello F, Riesterer O, and Sherweif M, et al (2007) Is EGFR a moving target during radiotherapy of carcinoma of the uterine cervix Gynecol Oncol 106 394–399 https://doi.org/10.1016/j.ygyno.2007.04.019 PMID: 17521713
56. Fuchs I, Vorsteher N, and Bühler H, et al (2007) The prognostic significance of human epidermal growth factor receptor correlations in squamous cell cervical carcinoma Anticancer Res 27 959–963 PMID: 17465227
57. Bertelsen BI, Steine SJ, and Sandvei R, et al (2006) Molecular Analysis of the PI3K-AKT pathway in uterine cervical neoplasia: frequent PIK3CA amplification and AKT phosphorylation Int J Cancer 118 1877–1883 https://doi.org/10.1002/ijc.21461
58. Kim GE, Kim YB, and Cho NH, et al (2004) Synchronous coexpression of epidermal growth factor receptor and cyclooxygenase-2 in carcinomas of the uterine cervix: a potential predictor of poor survival Clin Cancer Res 10 1366–1374 https://doi.org/10.1158/1078-0432.CCR-0497-03 PMID: 14977838
59. Cho NH, Kim YB, and Park TK, et al (2003) P63 and EGFR as prognostic predictors in stage IIB radiation-treated cervical squamous cell carcinoma Gynecol Oncol 91 346–353 https://doi.org/10.1016/S0090-8258(03)00504-3 PMID: 14599865
60. Ray A, Naik SLD, and Sharma BK (2002) Distribution of prognostically unfavourable product of c-erbB-2 oncogene and EGF-R in carcinomas of the breast and uterine cervix Indian J Physiol Pharmacol 46 423–433
61. Kim YT, Park SW, and Kim JW (2002) Correlation between expression of EGFR and the prognosis of patients with cervical carcinoma Gynecol Oncol 87 84–89 https://doi.org/10.1006/gyno.2002.6803 PMID: 12468347
62. Leung TW (2001) Expressions of c-erbR-2, epidermal growth factor receptor and pan-ras proto-oncogenes in adenocarcinoma of the cervix: correlation with clinical prognosis Oncol Rep 8 1159–1164 PMID: 11496335
63. Alonio LV, Dalbert D, and Picconi MA, et al (2000) Mutaciones en genes Ha-ras y p53 detectadas mediante PCR-SSCP en lesiones premalignas y malignas de cuello uterino asociadas con virus papiloma humano (HPV) Medicina 60 895–901
64. Ma Y-Y, Wei S-J, and Lin Y-C, et al (2000) PIK3CA as an oncogene in cervical cancer Oncogene 19 2739–2744 https://doi.org/10.1038/sj.onc.1203597 PMID: 10851074
65. Kim JW, Kim YT, and Kim DK (1990) Correlation between EGFR and c-erbB-2 oncoprotein status and response to neoadjuvant chemotherapy in cervical carcinoma Yonsei Med J 40 207–214 https://doi.org/10.3349/ymj.1999.40.3.207
66. Hove MG, Dinh TV, and Hannigan EV, et al (1999) Oncogene expression and microvessel count in recurrent and non-recurrent stage Ib squamous cell carcinoma of the cervix J Reprod Med 44 493–496 PMID: 10394542
67. Skomedal H, Kristensen GB, and Lie AK, et al (1999) Aberrant expression of the cell cycle associated proteins TP53, MDM2, p21, p27, cdk4, cyclin D1, RB, and EGFR in cervical carcinomas Gynecol Oncol 73 223–228 https://doi.org/10.1006/gyno.1999.5346 PMID: 10329038
68. Kersemaekers A-MF, Fleuren GJ, and Kenter GG, et al (1999) Oncogene alterations in carcinomas of the uterine cervix: Overexpression of the epidermal growth factor receptor is associated with poor prognosis Clin Cancer Res 5 577–586 PMID: 10100709
69. Biesterfeld S, Schuh S, and Muys L, et al (1999) Absence of epidermal growth factor receptor expression in squamous cell carcinoma of the uterine cervix is an indicator of limited tumor disease Oncol Rep 6 205–209
70. Parker MF, Arroyo GF, and Geradts J, et al (1997) Molecular characterization of adenocarcinoma of the cervix Gynecol Oncol 64 242–251 https://doi.org/10.1006/gyno.1996.4580 PMID: 9038270
71. Kristensen GB, Holm R, and Abeler VM, et al (1996) Evaluation of the prognostic significance of cathepsin D, epidermal growth factor receptor, and c-erbB-2 in early cervical squamous cell carcinoma: an immunohistochemical study Cancer 78 433–440 https://doi.org/10.1002/(SICI)1097-0142(19960801)78:3<433::AID-CNCR9>3.0.CO;2-K PMID: 8697388
72. Lee J-H, Lee S-K, and Yang M-H, et al (1996) Expression and mutation of H-ras in uterine cervical cancer Gynecol Oncol 62 49–54 https://doi.org/10.1006/gyno.1996.0188 PMID: 8690291
73. Tenti P, Romagnoli S, and Silini E, et al (1995) Analysis and clinical implications of K-ras gene mutations and infection with human papillomavirus types 16 and 18 in primary adenocarcinoma of the uterine cervix Int J Cancer 64 9–13 https://doi.org/10.1002/ijc.2910640104 PMID: 7665253
74. Hale RJ, Buckley CH, and Gullick WJ, et al (1993) Prognostic value of epidermal growth factor receptor expression in cervical carcinoma J Clin Pathol 46 149–153 https://doi.org/10.1136/jcp.46.2.149 PMID: 8459036 PMCID: 501147
75. Willis G, Jennings B, and Ball RY, et al (1993) Analysis of ras point mutations and human papillomavirus 16 and 18 in cervical carcinomata and their metastases Gynecol Oncol 49 359–364 https://doi.org/10.1006/gyno.1993.1140 PMID: 8390962
76. Koulos JP, Wright TC, and Mitchell MF, et al (1993) Relationships between c-Ki-ras mutations, HPV types, and prognostic indicators in invasive endocervical adenocarcinomas Gynecol Oncol 48 364–369 https://doi.org/10.1006/gyno.1993.1064 PMID: 8385060
77. Hayashi Y, Hachisuga T, and Iwasaka T, et al (1991) Expression of ras oncogene product and EGF receptor in cervical squamous cell carcinomas and its relationship to lymph node involvement Gynecol Oncol 40 147–151. https://doi.org/10.1016/0090-8258(91)90107-G PMID: 1707025
78. Sato S, Ito K, and Ozawa N, et al (1991) Expression of c-myc, epidermal growth factor receptor and c-erbB-2 in human endometrial carcinoma and cervical adenocarcinoma Tohoku J Exp Med 165 137–145 https://doi.org/10.1620/tjem.165.137 PMID: 1687493
79. De La Rochefordiere A, Kamal M, and Floquet A, et al (2015) PIK3CA pathway mutations predictive of poor response following standard radiochemotherapy ± cetuximab in cervical cancer patients Clin Cancer Res 21 2530–2537 https://doi.org/10.1158/1078-0432.CCR-14-2368 PMID: 25724520
80. Yamashita H, Murakami N, and Asari T, et al (2009) Correlation among six biologic factors (p53, p21WAF1, MIB-1, EGFR, HER2, and Bcl-2) and clinical outcomes after curative chemoradiation therapy in squamous cell cervical cancer Int J Radiat Oncol Biol Phys 74 1165–1172 https://doi.org/10.1016/j.ijrobp.2008.09.005
81. Oka K, Nakano T, and Arai T (1997) Expression of cathepsin D and epidermal growth factor receptor in stage III cervical carcinomas Int J Gynecol Cancer 7 122–126 https://doi.org/10.1046/j.1525-1438.1997.00431.x
82. Wistuba I, Roa I, and Araya J, et al (1994) Immunohistochemical expression of epidermal growth factor receptor (EGFR) in epidermoid carcinoma of the cervix uteri and its precursor lesions Rev Chil Obstet Ginecol 59 116–122
83. Bauknecht T, Kohler M, and Janz I, et al (1989) The occurrence of epidermal growth factor receptors and the characterization of EGF-like factors in human ovarian, endometrial, cervical and breast cancer - EGF receptors and factors in gynecological carcinomas J Cancer Res Clin Oncol 115 193–199 https://doi.org/10.1007/BF00397923
84. Wistuba OI and Capurro VI (1995) Mutaciones del gen K-ras y expresión inmunohistoquímica de la proteína ras-p21 en cáncer de cuello uterino Rev Chil Obstet Ginecol 60 282–287
85. Lachkar B, Minaguchi T, and Akiyama A, et al (2018) Prognostic significance of PIK3CA mutation in stage IIB to IVA cervical cancers treated by concurrent chemoradiotherapy with weekly cisplatin Medicine 97(31) e11392 https://doi.org/10.1097/MD.0000000000011392 PMID: 30075505 PMCID: 6081058
86. Jiang W, Xiang L, and Pei X, et al (2018) Mutational analysis of KRAS and its clinical implications in cervical cancer patients J Gynecol Oncol 29 e4 https://doi.org/10.3802/jgo.2018.29.e4
87. Spaans VM, Nyoman Bayu Mahendra I, and Purwoto G, et al (2018) The landscape of somatic mutations in Indonesian cervical cancer is predominated by the PI3K pathway Gynecol Oncol 148 189–196 https://doi.org/10.1016/j.ygyno.2017.10.009
88. de Almeida VH, de Melo AC, and Meira DD, et al (2017) Radiotherapy modulates expression of EGFR, ERCC1 and p53 in cervical cancer Braz J Med Biol Res 51 e6822
89. Hodgson A, Amemiya Y, and Seth A, et al (2017) Genomic abnormalities in invasive endocervical adenocarcinoma correlate with pattern of invasion: biologic and clinical implications Mod Pathol 30 1633–1641 https://doi.org/10.1038/modpathol.2017.80 PMID: 28731050
90. Wei H, Wang X-W, and Chen K-M, et al (2018) Analysis of gene mutation associated with tyrosine kinase inhibitor sensitivity of epidermal growth factor receptor in cervical cancer patients Eur Rev Med Pharmacol Sci 22 6280–6287 PMID: 30338793
91. Zou Y, Liu FY, and Wu J, et al (2017) Mutational analysis of the RAS/RAF/MEK/ERK signaling pathway in 260 Han Chinese patients with cervical carcinoma Oncol Lett 14 2427–2431 https://doi.org/10.3892/ol.2017.6435 PMID: 28781678 PMCID: 5530172
92. Ueda A, Takasawa A, and Akimoto T, et al (2017) Prognostic significance of the co-expression of EGFR and HER2 in adenocarcinoma of the uterine cervix PLoS One 12 e0184123 https://doi.org/10.1371/journal.pone.0184123 PMID: 28859123 PMCID: 5578660
93. Borges Migliavaca C, Stein C, and Colpani V, et al (2020) How are systematic reviews of prevalence conducted? A methodological study BMC Med Res Methodol 20 96 https://doi.org/10.1186/s12874-020-00975-3 PMID: 32336279 PMCID: 7184711
Appendix A. Search strategy.
Database: Ovid MEDLINE(R), Epub Ahead of Print, In-Process and Other Non-Indexed Citations, Ovid MEDLINE(R) Daily and Ovid MEDLINE(R) <1946 to Present>
Search strategy:
-------------------------------------------------------------------------------------------------------------------------------------------------------------------
1) exp Uterine Cervical Neoplasms/
2) (cervi* adj5 (cancer* or tumo?r* or neoplas* or carcinoma* or malignan* or adenocarcinoma* or choriocarcinoma* or teratoma* or sarcoma*)).ti,ab.
3) 1 or 2
4) exp Phosphatidylinositol 3-Kinases/
5) (Phosphatidylinositol 3 adj1 kinas*).ti,ab.
6) (PI3K* or PI 3K* or PI-3K* or PI 3-kinase* or PI3 kinase*).ti,ab.
7) (ptdlns adj3 kinase*).ti,ab.
8) phosphatidylinositol-3-oh kinase*.ti,ab.
9) 4 or 5 or 6 or 7 or 8
10) exp Genes, ras/
11) ras.ti,ab.
12) 10 or 11
13) exp Proto-Oncogene Proteins B-raf/
14) raf.ti,ab.
15) 13 or 14
16) kras protein, human.mp.
17) kras.ti,ab.
18) 16 or 17
19) exp Receptor Protein-Tyrosine Kinases/
20) (receptor* adj3 kinase*).ti,ab.
21) 19 or 20
22) exp Receptor, Epidermal Growth Factor/
23) (epiderm* adj3 factor*).ti,ab.
24) 22 or 23
25) exp ErbB Receptors/
26) (erbb adj3 receptor*).ti,ab.
27) 25 or 26
28) exp Genetic Variation/
29) (genet* adj3 (mutat* or variat*)).ti,ab.
30) (therap* adj3 target*).ti,ab.
31) 28 or 29 or 30
32) 9 or 12 or 15 or 18 or 21 or 24 or 27 or 31
33) 3 and 32
34) 9 or 12 or 15 or 18 or 21 or 24 or 27
35) 3 and 34
***************************
Appendix B. Excluded studies.
The following table gathers the references of the studies that were excluded (N = 93) at the full text review stage. They are grouped according to reason for exclusion.
Appendix C. Study methodology evaluation tool used in this review.

Appendix D. Figures.
EGFR
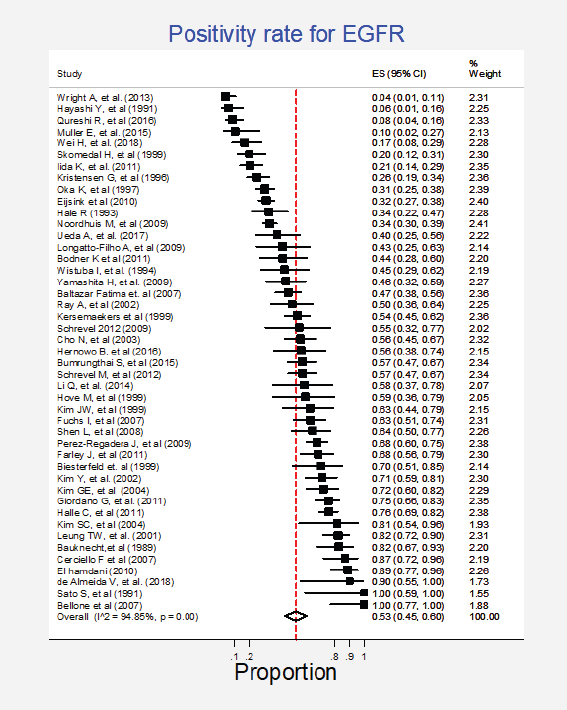
Figure 1. Meta-analysis of the EGFR positivity rates.
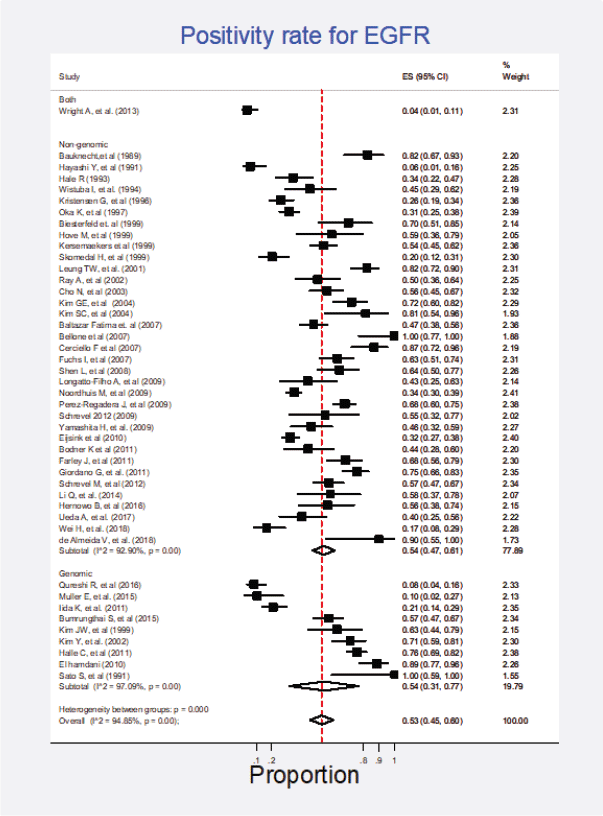
Figure 2. Subgroup analysis. Positivity for EGFR according to the type of detection test (genomic versus non-genomic).
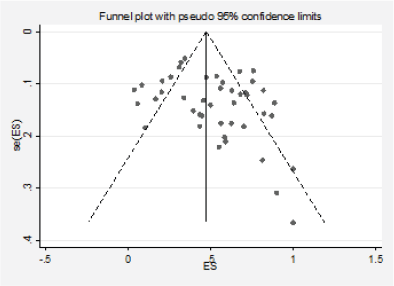
Figure 3. Analysis of the effect of small studies on the EGFR positivity rate (assessment of publication bias).
PI3K
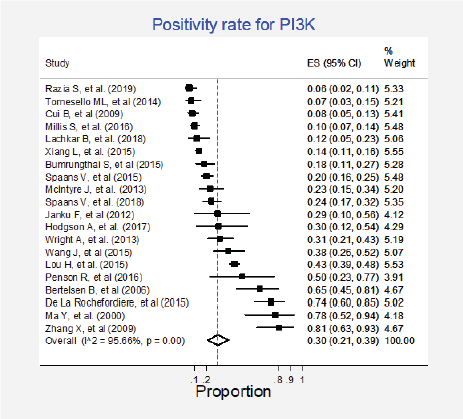
Figure 4. Meta-analysis of the PI3K positivity ratios.
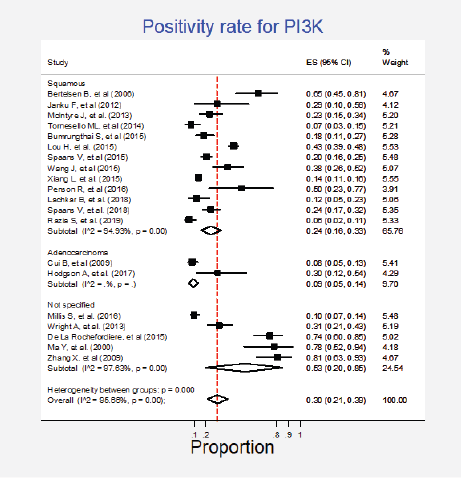
Figure 5. Subgroup analysis. Positivity for PI3K according to the type of histology.
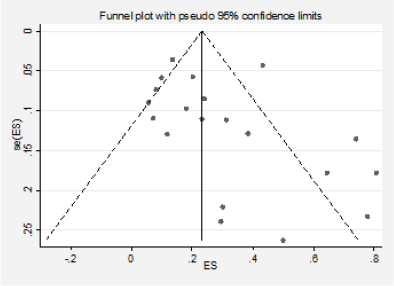
Figure 6. Analysis of the effect of small studies on the PI3K positivity rate (assessment of publication bias).
Ras
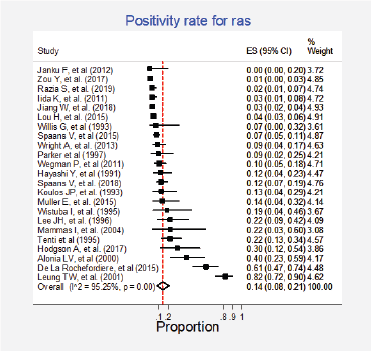
Figure 7. Meta-analysis of the Ras positivity proportions.
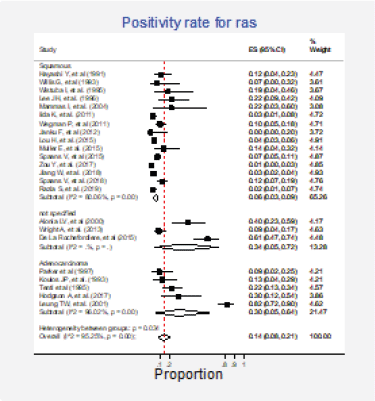
Figure 8. Subgroup analysis. Positivity for Ras according to the histological type.
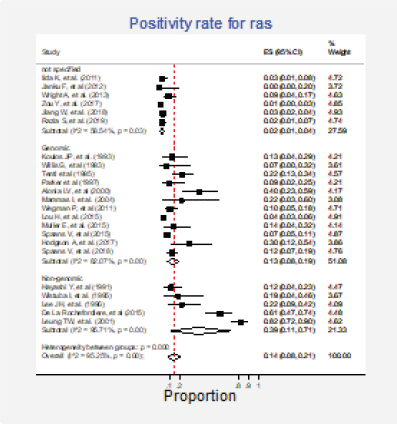
Figure 9. Subgroup analysis. Positivity for Ras according to the type of detection test (genomic versus non-genomic).
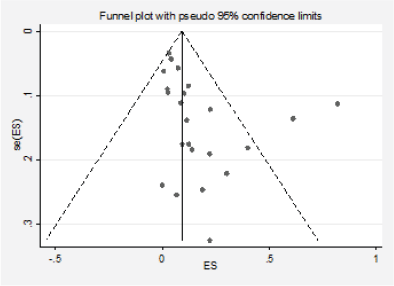
Figure 10. Analysis of the effect of small studies on the Ras positivity rate (assessment of publication bias).
Akt
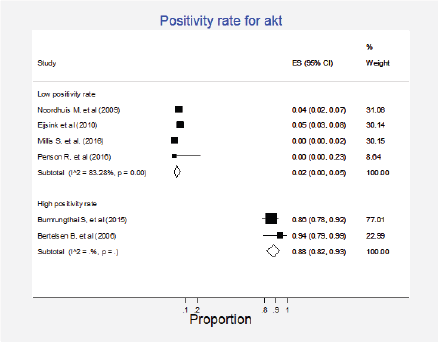
Figure 11. Meta-analysis of the positivity proportions of Akt.
Raf
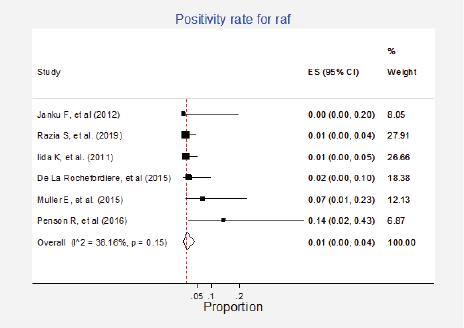
Figure 12. Meta-analysis of the Raf positivity ratios.